OEToolkits 2017.Jun
Shape TK: Hermite
The 2017.Jun release introduces the OEHermite class, which allows a molecule to be expanded into a Hermite representation. An initial suggested area of application is the representation of protein shape. The advantages of representing proteins by Hermites include:
smooth representations that capture any level of detail, from the atomistic (many coefficients, see the picture on the right) to the very coarse (few coefficients, as in the middle picture),
the ability to store these representations in a compact manner,
the ability to transform these representations easily, and
very fast overlap calculations between proteins.
Below are examples of low- and high-resolution Hermite representation images of dihydrofolate reductase (DHFR):
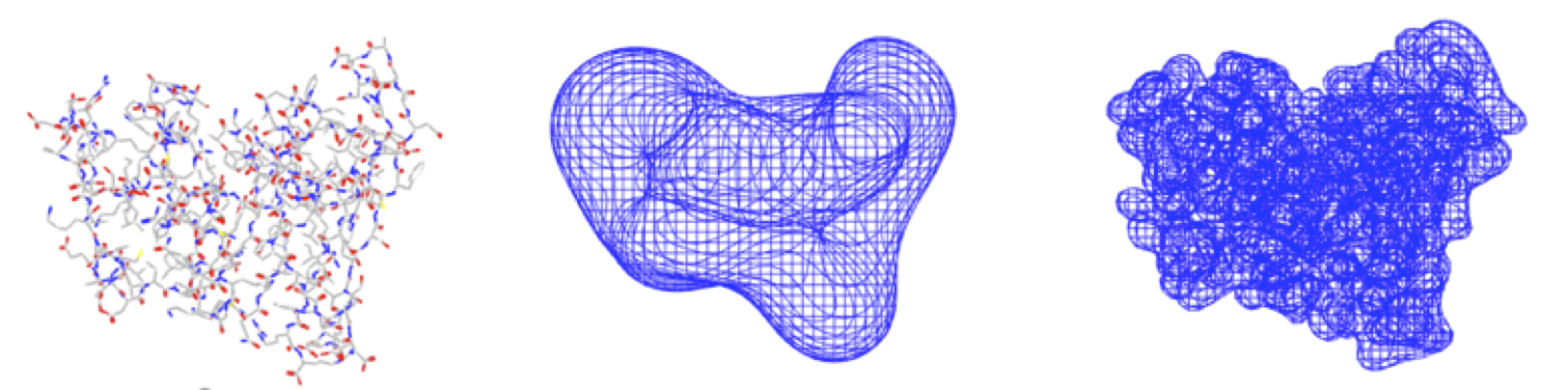
Exact shape versus Hermite Representation
OpenEye will explore the potential of Hermite representations; we invite users to do likewise with this exciting new addition to the OpenEye toolkits. See the Shape from Hermite Representation chapter of the Shape TK documentation for more information.
OEDepict TK and Grapheme TK: Interactive SVG images
Scalable Vector Graphics (SVG) image format is dominant in web applications due to its wide browser support and scalability. An SVG image contains a list of graphical objects that can be grouped, styled, and transformed together. This enables the generation of interactive images defined in the SVG XML elements or via scripting that accesses the SVG Document Object Model (DOM).
The new Preliminary API available in OEDepict TK provides low-level functionality that enables grouping of graphical elements in SVG images that can be manipulated externally by web developers. High-level API entrypoints are also available to generate self-contained SVG images that show or hide different elements of the image based on mouse events.
Hover the mouse over any ligand atom to display its b-factor |
Click on any residue circle to display ligand atoms it interacts with |
|
|
For more information see the Generating Interactive SVG Images chapter of the OEDepict TK documentation.
GraphSim TK: Fast Fingerprint Similarity Search
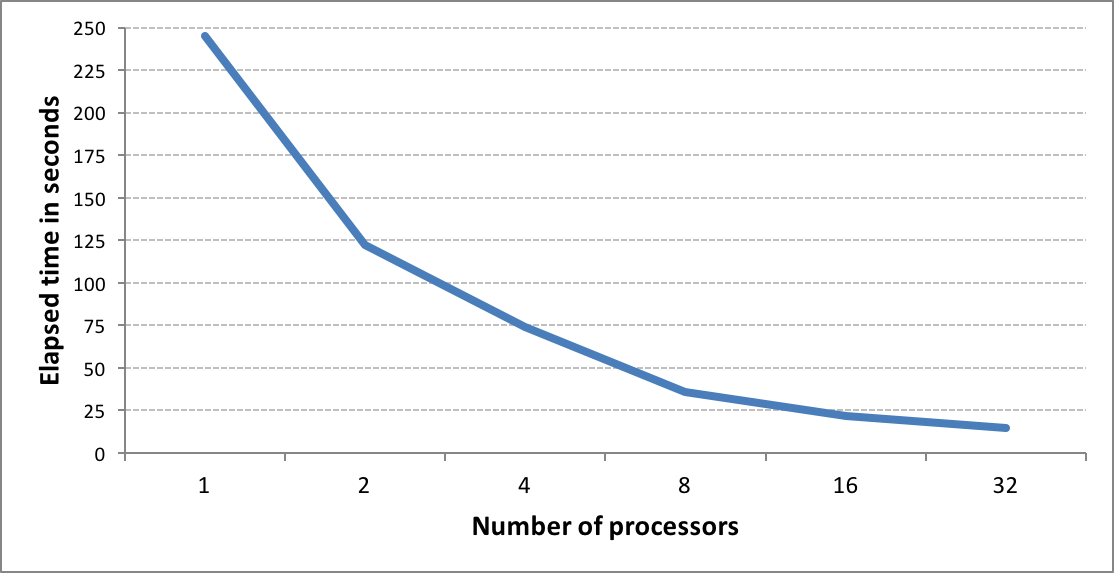
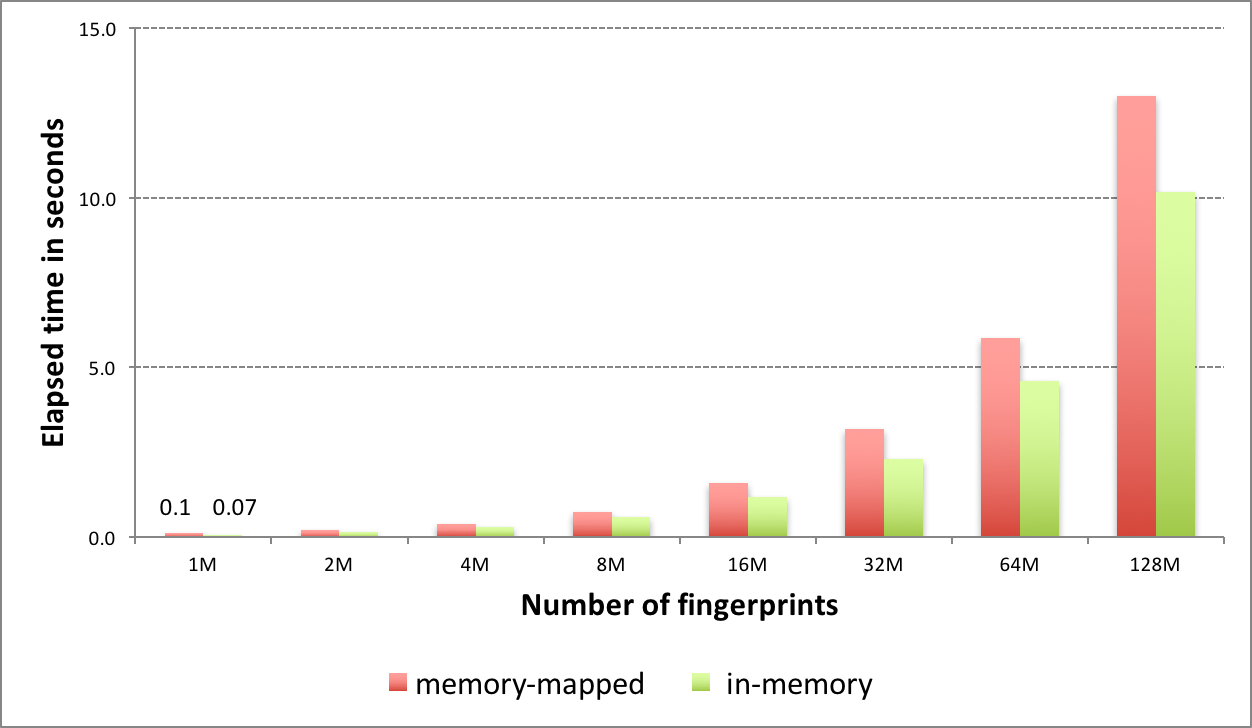
OEGraphSim TK now provides improved fingerprint search capability, allowing users to perform molecule similarity searches on large databases up to 10x faster than previous versions.
Fingerprint methods are used for the computation of similarity scores. When searching large virtual databases, performance can be problematic. To address this issue, OEGraphSim TK now uses popcount, a hardware instruction available on modern CPUs, to provide significant performance improvements for computing similarity.
The new fingerprint search can be performed in two modes: in-memory and memory-mapped. The in-memory mode involves preloading all fingerprints into memory and performing the search. While this represents the fastest way to perform similarity searches once the fingerprints are loaded, searches are limited by memory availability. The memory-mapped mode has no load time penalty or memory limitation but the search itself takes more time.
See the GraphSim TK Examples chapter of the OEGraphSim TK documentation for more information and examples using the new OEFastFPDatabase class.
General Notices
On Linux, Python
single-builddistributions are now installed by default when using the meta-installer viapip install openeye-toolkits. If a problem is encountered with thesingle-buildpackages, users can set the environment variableOE_PIP_ARCH=oldbefore thepip installstep.Python 3.6 is supported in the 2017.Jun release.
This is the last release to support 32-bit Java.
This is the last release to support Red Hat Enterprise Linux version 5. As of March 31, 2017, RHEL5 has reached end-of-production support.
Note
OpenEye continues to monitor and collect feedback from our customers on the need to support Python 2 wrappers. Our current plan is to phase out support for Python 2 on all platforms by the 2017.Oct release. As this is a substantial change for our customers, we are willing to help with migration. Please contact support@eyesopen.com for more details.