FastROCS Architecture
Memory Usage
A rule of thumb is 1 GB of main memory for every 4 million conformers. The primary variable FastROCS TK users need to think about is how many conformers per molecule is needed. The OMEGA default of 200 conformers per molecule is much higher than it needs to be due to OEDocking vs ROCS virtual screening performance. OEDocking needs a higher number of conformers than ROCS does. ROCS can perform quite well with 10-50 conformers per molecule. The number of conformers chosen has a direct linear relationship to FastROCS query performance and the amount of memory needed.
Note
The “rule of thumb” is based upon the number of heavy atoms being normally distributed around 25-30 heavy atoms. The number of heavy atoms also has a linear relationship to the memory consumption. So fragment databases will be significantly smaller, but large macro-cycle peptide databases will be significantly larger.
Note
The above “rule of thumb” assumes the default of four inertial starts and no alternative starting points. If you choose to use eight inertial starts, you should assume you’ll need about 1.5 as much main memory, i.e. 1.5 GB for every 4 million conformers. If you choose to use alternative starts, you should multiply the amount of main memory by the number of alternative starts / 3.
Color scoring does significantly increase virtual screening
performance. As much color information, centroids and self terms, is
cached in memory as well. For doing pure
OEShapeDatabaseType_Shape analysis, color
information can be elided allowing for a small percentage in memory
size reduction, around 5-10%. For a more complete breakdown of memory
usage, use the
OEShapeDatabase.PrintMemoryUsage method to
print statistics on memory usage.
Note
Color atom and self term calculation is a dominating factor in
how fast FastROCS can load molecules from disk. Therefore, it
is heavily recommended to pre-process OEB files with
OEPrepareFastROCSMol.
It is important to note that FastROCS is most sensitive to host main memory, not GPU memory. On board GPU memory size does not heavily affect FastROCS performance, so buying the cheaper GPUs with less memory is just fine for FastROCS.
Note
This memory overhead will be improved over time. Significant advances were made in version 1.4.0, memory consumption per conformer was cut in half. Then cut in half again for the 1.7.0 release.
Multi-GPU Scaling
FastROCS will automatically scale across as many GPUs as can be found on the current machine. It is not uncommon or overly expensive to get a machine with 4-8 GPUs in it. As with any parallel program, truly optimal linear scaling is difficult to achieve, but FastROCS gets fairly close as can be seen in the below graph.
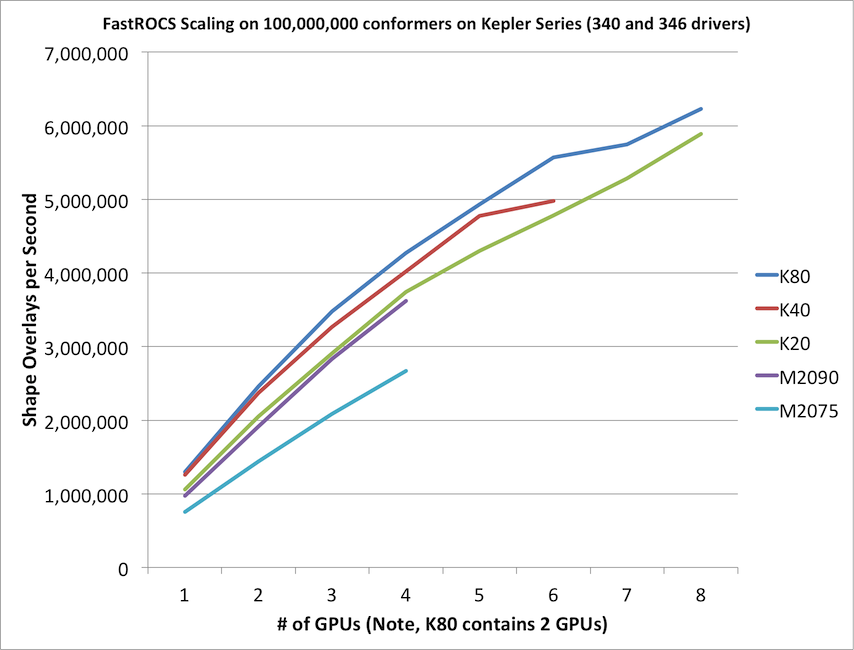
This graph is a comparison of the various generations of Tesla compute cards produced by NVidia for high performance computing purposes.
Note
Static color calculations are still performed on the CPU. For best performance, a decent mid-range CPU with 2-cores per GPU is recommended.
The OEShapeDatabase class will by default automatically scale across all GPUs and CPUs in the system. In this way it can be a very large resource hog for CPU, GPU, and memory. There are some methods provided on the class to control resource utilization, but the current best control is through the CUDA_VISIBLE_DEVICES environment variable.
Multi-Node Scaling
Scaling FastROCS across multiple nodes is possible with the included
ShapeDatabaseProxy.py example.