OESubSearchDatabase
Attention
This API is currently available in C++ and Python.
class OESubSearchDatabase
The OESubSearchDatabase class is designed to perform fast multi-threaded substructure search. Each OESubSearchDatabase is associated with one screen type (OESubSearchScreenTypeBase) that is set when the database is initialized from a pre-generated binary substructure search file.
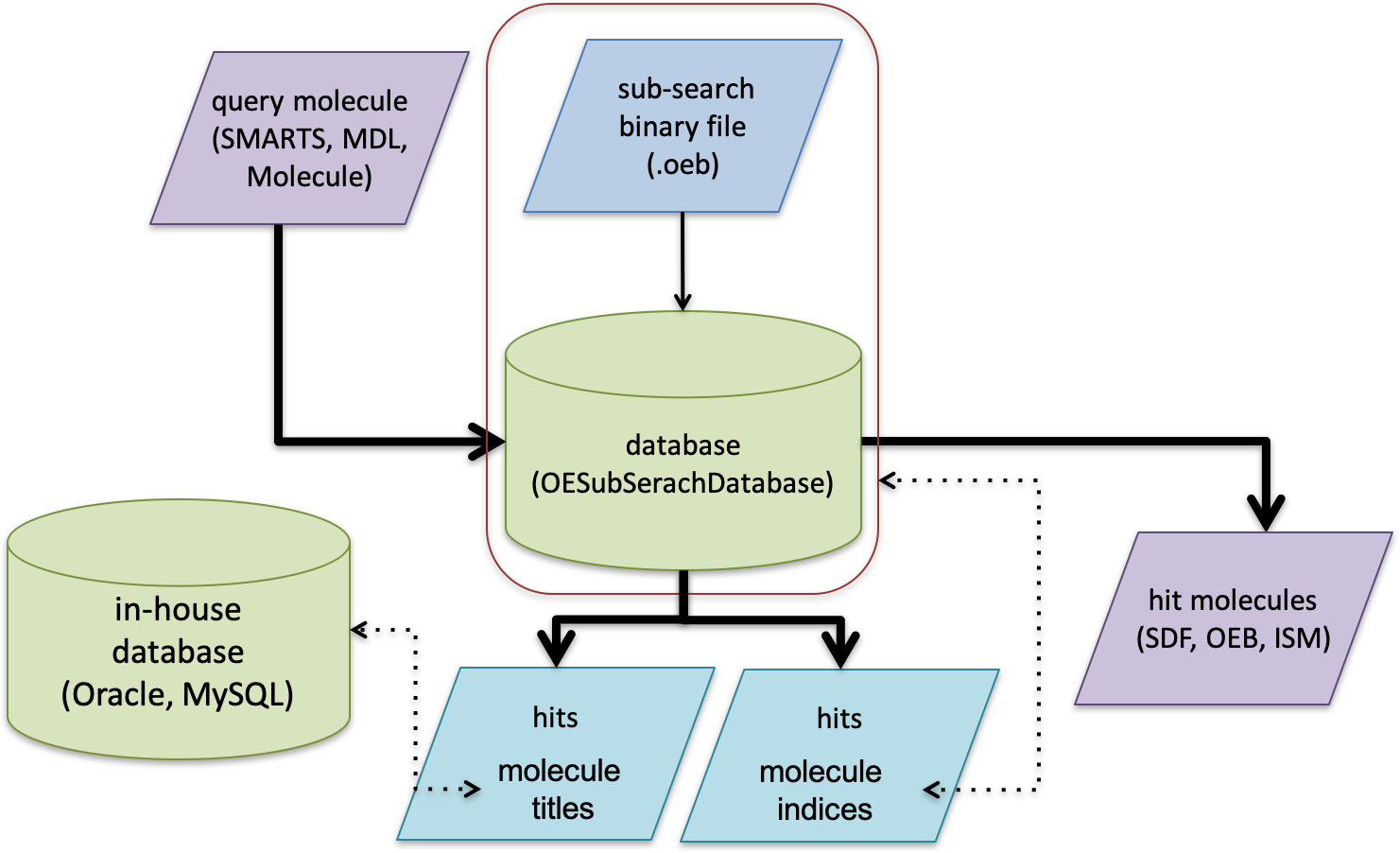
Schematic representation of using substructure search database
The OESubSearchDatabase class provides the following search methods:
Method |
Returns |
Type |
|---|---|---|
number of matched molecules |
full search |
|
indices of matched molecules |
limited search |
|
titles of matched molecules |
limited search |
|
|
full/limited search (cancelable) |
See also
OECreateSubSearchDatabaseFilefunction to generate database files for OESubSearchDatabase.
Code Example
Constructors
OESubSearchDatabase(const std::string &dbfile,
const unsigned memtype=OESubSearchDatabaseType::Default,
const unsigned numprocesssors=0u)
OESubSearchDatabase(const unsigned memtype=OESubSearchDatabaseType::Default,
const unsigned numprocesssors=0u)
Constructs an OESubSearchDatabase object.
- dbfile
The name of the file that contains the substructure search data (molecules with pre-generated screens). The file has to be generated with the
OECreateSubSearchDatabaseFilefunction. If no file name is defined, the database has to be initialized by calling theOESubSearchDatabase::Openmethod.- memtype
Defines the type of the substructure database. This value has to be from the
OESubSearchDatabaseTypenamespace.- numprocesssors
The maximum number of threads that can be used to search the database. When set to
0, the number of processors used will be the number returned by theOEGetNumProcessorsfunction. Ifnumprocesssors == 1, all searches are executed using the main thread only.
GetDatabaseType
unsigned GetDatabaseType() const
Returns the type of the substructure search database.
The return value is taken from the OESubSearchDatabaseType
namespace.
GetMatchIndices
OESystem::OEIterBase<size_t>* GetMatchIndices(const OESubSearchQuery &) const
Returns the indices of molecules matched to the given query
(OESubSearchQuery).
The search terminates if the limit set for the query is reached
(GetMaxMatches).
The matched molecules can be retried from the database by using the
OESubSearchDatabase::GetMolecule method.
GetMatchTitles
OESystem::OEIterBase<std::string>* GetMatchTitles(const OESubSearchQuery &) const
Returns the titles of the molecules matched to the given query
(OESubSearchQuery).
The search terminates if the limit set for the query is reached
(GetMaxMatches).
GetMolecule
bool GetMolecule(OEMolBase &mol, const size_t index) const
Retrieves the molecule stored in the database with the specified index.
See also
GetScreenType
const OESubSearchScreenTypeBase *GetScreenType() const
Returns the screen type of the OESubSearchDatabase object. An OESubSearchDatabase object can only store screens with identical types.
GetTitle
std::string GetTitle(const size_t index) const
Retrieves the title of the molecule stored in the database with the specified index.
See also
IsValid
bool IsValid() const
Returns whether the OESubSearchDatabase object is valid.
NumMatches
size_t NumMatches(const OEQMolBase &query) const
Returns the number of molecules matched to the given query (OESubSearchQuery) in the entire database.
NumMolecules
size_t NumMolecules() const
Returns the number of molecules stored in the OESubSearchDatabase object.
NumProcessors
unsigned NumProcessors() const
Returns the maximum number of threads that can be used to search the database. This is determined when the database is initialized.
Open
bool Open(oemolistream &ifs,
OESystem::OETracerBase &tracer=OESystem::OENoTracer)
bool Open(const std::string & filename,
OESystem::OETracerBase &tracer=OESystem::OENoTracer)
Initializes the substructure search database. These methods return
true if the database was successfully initialized.
- ifs
The molecule stream from which the substructure search database is initialized.
- filename
The name of the file from which the substructure search database is initialized.
- tracer
The tracer that can be used to report the progress of the database load.
See also
OEConsoleProgressTracer class
Search
unsigned Search(OESubSearchResult &result, const OESubSearchQuery &query) const
This search method is designed for interactive searches.
- result
The OESubSearchResult object that stores the result of the search along with the progress of the search. The OESubSearchResult is updated regularly during the search as more and more matches identified. It also provides a mechanism to cancel a long running search via
OESubSearchResult::Cancel.- query
The OESubSearchQuery object that stores the query molecule with search parameters.
The OESubSearchDatabase::Search method returns:
OESubSearchStatus::Uninitializedif the search can not be executedOESubSearchStatus::Finished, if the search successfully finishedOESubSearchStatus::Canceled, if the search was interrupted
See also
OESubSearchStatusnamespace