Version 2.0.3
OEChem 2.0.3
New features
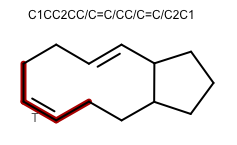
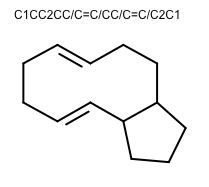
OEGenerate2DCoordinatesadded the following new features to improve the 2D layout of molecules:The ring template dictionary search algorithm now has the ability to preserve cis/trans bond stereo. The 2014.Feb and 2014.Jun releases contained a bug that caused bond stereo to be erroneously inverted. The 2014.Oct release prevented the erroneous inversion by disallowing creation of such 2D coordinate layouts. This release is the next step in improving 2D coordinate generation for large macrocycles: ring templates with matching cis/trans stereo chemistry can now be used.
252 new ring templates have been added:
180 complex ring systems with anonymized carbon frameworks
72 common macrocycle ring systems with specified cis/trans bond configuration
Example of 2D coordinate generation for macrocycles with cis/trans bond stereo OEChem TK 2.0.2
OEChem TK 2.0.3
A new OEExtractRingTemplateOptions class and
OEExtractRingTemplatesfunction, which are used to extract potential new ring templates, have been added.A new OEChem TK example that extracts ring templates for 2D coordinate generation has been added. For more information, see the Extract 2D ring templates section.
Note
To improve the depiction of the ring systems in a collection of molecules, please run the Extract 2D ring templates script on the collection and send the results to support@eyesopen.com. OpenEye will be happy to include the ring systems in the next release of the toolkits.
A new
OEIs2DFormatfunction that can determine whether the given file format supports 2D coordinate information has been added.Canonical SMILES generated with the
OESMILESFlag::SuperAtomsflag will now respect any value that is placed in theOEAtomBase::GetNamefield while canonical ordering the molecule. This feature is useful for creating specialized SMILES that can be canonicalized and deduplicated.A new
OEReadOEBFilereader has been added that makes low-level OEB parsing available to Python, Java, and C#. Note: this function is considered advanced;OEReadMoleculeis still preferred.
Major bug fixes
InChI creation will now fail if the molecule being converted cannot be handled by the InChI reference implementation. The OEChem TK will throw a warning message about the molecular data that cannot be converted and return an empty string. This also protects against many possible crashes inside the InChI reference implementation due to abnormal molecules.
InChI creation will no longer overflow the stack when an atom has more than 20 neighbors.
The state of the
chiralflag from MDL files is now correctly managed and maintained byOEReadMoleculeandOEWriteMolecule. Previously,non-chiral(relative) stereochemistry structures were being promoted incorrectly tochiral(absolute) stereochemistry.OESubSearch constructors no longer create or use uninitialized memory. This bug was introduced in the 2014.Jun release and caused erroneous warning messages about explicit and implicit hydrogen mismatches.
OEPDBOrderAtomspreviously sorted residues improperly due to a buffer overflow for theOEResidueIndex::YGresidue index corresponding to the T-RNA base wybutosine. This has been fixed.OEWritePDBFilewill no longer buffer overflow for atoms that have their name fields set.OEWriteMacroModelFilewill no longer buffer overflow for atoms that have their name fields set.OELibraryGen previously had a thread-safety issue when generating libraries. This has been fixed.
Java-specific changes
OEGetSDDataPairswill again return aOESDDataIter. The return value changed in the 2014.Jun release from “OESDDataIter” to “OEPDBDataIter” by accident.
API changes
The following public API points were largely undocumented and have been removed as internal implementation details, including the removal of the public header:
OEFuzzy OEGetAtomicNumconst OEExprBase*,...OEFuzzy OEGetAromatic(const OEExprBase*,...)OEFuzzy OEGetFormalCharge(const OEExprBase*,...)OEFuzzy OEGetImplicitHCount(const OEExprBase*,...)OEFuzzy OEGetHCount(const OEExprBase *expr,...)OEFuzzy OEGetBondOrder(const OEExprBase*,...)bool OEIsSingleOrAromatic(const OEExprBase*)OEFuzzy OEGetStereo(const OEExprBase *expr,...)
Additionally, the function
OEDisassembleExpressionshas been explicitly marked as deprecated in the documentation and will be removed in a future release.
Minor bug fixes
OEMolBase::Clearwill no longer leak memory forOEMCMolType::OEDBMCMolafterOEMolBase::Compresshas been called.A warning is now thrown if kekulization is skipped and
OEParseSmilessubsequently finds a bond with a bond order of five.Warnings will now be issued for enhanced stereo collection in V3000 format files that have atoms in more than one enhanced stereo collection. A given stereo center atom can be in no more than one stereo collection type.
Enhanced stereo collection information in V3000 MDL files is now correctly parsed for multiline appendix information.
Parsing of V3000 stereo collection information has been corrected to combine explicitly named stereo collections. Previously, multiple stereo collection entries of the same name in separate appendix entries generated different stereo groups.
For V3000 MDL files, implicitly defined stereo collection information is now explicitly exported from
OEWriteMolecule. Defined stereocenters that are not included in the explicit enhanced stereo collections will be added to the appropriate stereo collection based on the state of thechiralflag.For MDL files, the writer will always attempt to export in V2000 format to ensure maximum compatibility with third party programs. If V3000 format files are explicitly desired regardless of whether the molecule is compatible with V2000, use
OEOFlavor::MDL::MV30.Undefined behavior in
OEMolBaseType::OEMiniMolhas been fixed.OEMolDatabase::Savewill now throw a warning message and returnfalsewhen attempting to save the database to the same file.OEMCMolBase::NewConfwill now throw a warning message if the number of atoms in the OEMolBase or OEConfBase is different than the OEMCMolBase into which the conformer is being constructed. This addresses the common problem of creating conformers from molecules that simply differ in the number of implicit or explicit hydrogens. Note:OEMCMolBase::NewConfis still designed to be very fast, so the safety checks are intentionally kept minimal.OESetRotamernow properly returnstrueto indicate success when applied to residues that have fewer than four chi angles.
Documentation fixes
Extensive documentation has been added to the
OEExprOptsnamespace and how it affects OESubSearch and OEMCSSearch.An example code has been added that describes how to retrieve an OEGraphMol object as a OEInterface parameter.
A large warning message has been added to the Multi-Threaded OEChem chapter, alerting users that passing molecules between threads is not supported by default.
OEBio 2.0.3
New features
A new OEBio::OEFragmentNetwork class that represents networks of fragments and their interactions has been added. This is being used by the new protein-ligand rendering capability in the Grapheme TK. This class is considered beta and will likely change over the next year as the protein-ligand rendering feature is refined.
OESystem 2.0.3
New features
OENumberToStringwill now acceptlong doublearguments.OEIsSafeCasthas been added for determining whether a given value can be safely cast to a particular return type without loss of information.
Minor bug fixes
OESystem::OEBinaryReadno longer crashes nor displays undefined behavior when there is a mismatch between the template parameter and the registered reader objects.OEUniMolecularRxn now strips internal generic atom data annotations on the generated product molecules.
OEUniMolecularRxn now corrects implicit hydrogen counts on charged atoms in the product molecules.
Documentation fixes
Function documentation for
OEParseHexhas been added.
OEPlatform 2.0.3
New features
OEGetPlatformInfowill now generate more accurate descriptions of modern Windows operating systems.
Minor bug fixes
OEGetHostIdentwill no longer leak memory.
Documentation fixes
Documentation has been added for the following functions:
OEGrid 1.5.0
New features
OEReadMTZwill now automatically recognize column names generated by PHENIX in MTZ files.