InertialAtHeavyAtoms
OEFastROCSOrientation.InertialAtHeavyAtoms translates
the center of mass (COM) of each database molecule to each heavy atom of the
query molecule and to the query molecule’s COM. 4 inertial orientations are
then optimized at each translation.
The figures below demonstrate the starting positions optimized for the first 2 heavy atoms in the query molecule:
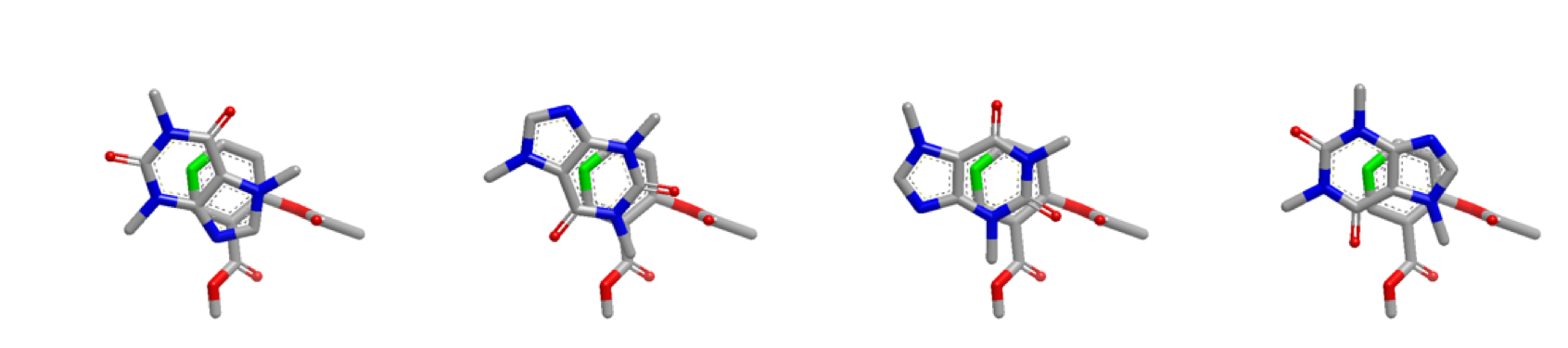
Inertial starts optimized at heavy atom 1 (highlighted green)
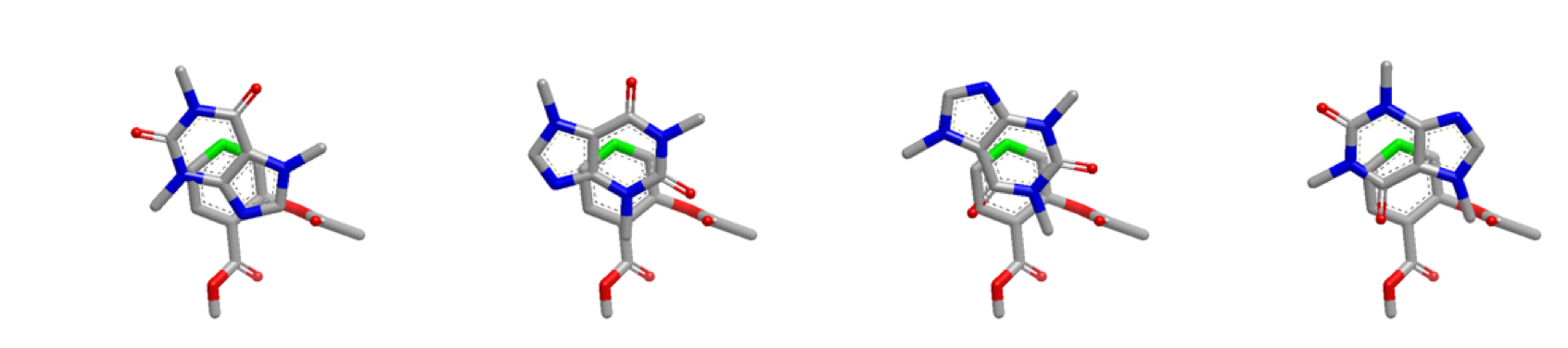
Inertial starts optimized at heavy atom 2 (highlighted green)
Inertial starts continue to be optimized for the remaining atoms of the query molecule and it’s COM.
Below is a link to a python script demonstrating the most basic usage of FastROCS TK.
After explaining this simple script it will be modified to make use of the
OEFastROCSOrientation.InertialAtHeavyAtoms
function.
Download code
The script creates an OEMolDatabase and an
OEShapeDatabase object. The database file is first
opened as an OEMolDatabase object, which is used to load
the molecules into OEShapeDatabase via
OEShapeDatabase.Open. This is the simplest example
of loading a database. The database is now ready for either combo or
shape-only searches.
Note
To restrict searches to shape-only, create a shape-only database object:
shapeOnlyDB = oefastrocs.OEShapeDatabase(oefastrocs.OEShapeDatabaseType_Shape)
While this reduces the memory footprint of the database as color data is no longer stored, the database cannot be searched for color scores.
In this example, an OEShapeDatabaseOptions object is
also created and the number of results is limited to 5 via the
OEShapeDatabaseOptions.SetLimit method.
Next, the query molecule is set up is using a molecule stream and the
OEReadMolecule function. Queries are used to search the
loaded database via the OEShapeDatabase.GetSortedScores
method. The OEShapeDatabaseOptions class is passed to
customize the search options. In this example, a subset of the eMolecules
database was searched against a benzene query molecule.
The top 5 scores are shown below.
Opening database file eMolecules.2015.1_100subset.oeb ...
Searching for benzene.oeb
Score for mol 65(conf 1) 0.530182 shape 0.237350 color
Score for mol 45(conf 1) 0.678128 shape 0.085376 color
Score for mol 49(conf 1) 0.596801 shape 0.153465 color
Score for mol 92(conf 0) 0.503204 shape 0.155330 color
Score for mol 33(conf 1) 0.442559 shape 0.175599 color
To make use of the
OEFastROCSOrientation.InertialAtHeavyAtoms
feature, modify the OEShapeDatabaseOptions object
as follows:
opts = oefastrocs.OEShapeDatabaseOptions() opts.SetLimit(5) opts.SetInitialOrientation(oefastrocs.OEFastROCSOrientation_InertialAtHeavyAtoms)
An additional option,
OEShapeDatabaseOptions.SetInitialOrientation, is
also set to the desired method,
OEFastROCSOrientation.InertialAtHeavyAtoms.
The resultant scores reflect the change in starting points:
Opening database file eMolecules.2015.1_100subset.oeb ...
Searching for benzene.oeb
Score for mol 45(conf 1) 0.669345 shape 0.121420 color
Score for mol 65(conf 1) 0.530182 shape 0.237350 color
Score for mol 49(conf 1) 0.596801 shape 0.153465 color
Score for mol 92(conf 0) 0.503204 shape 0.155330 color
Score for mol 33(conf 1) 0.442559 shape 0.175599 color
The OEShapeDatabaseOptions.GetNumHeavyAtomStarts
method can be used to
find out how many heavy atom starts are being optimized.
This method requires the query molecule in order to return the number of starts:
query = oechem.OEGraphMol() oechem.OEReadMolecule(qfs, query) if opts.GetInitialOrientation() == oefastrocs.OEFastROCSOrientation_InertialAtHeavyAtoms: numStarts = opts.GetNumHeavyAtomStarts(query) oechem.OEThrow.Info("This example will use %u starts" % numStarts)
When this code snippet is added to the Python script, the output should look like this:
This example will use 7 starts
Searching for benzene.oeb
Score for mol 45(conf 1) 0.669345 shape 0.121420 color
Score for mol 65(conf 1) 0.530182 shape 0.237350 color
Score for mol 49(conf 1) 0.596801 shape 0.153465 color
Score for mol 92(conf 0) 0.503204 shape 0.155330 color
Score for mol 33(conf 1) 0.442559 shape 0.175599 color
Download code
The fully modified python script used in this tutorial can be found here
InertialAtHeavyAtomStartsExample.py