Tutorial 2: How to prepare a database for faster load speeds
In this tutorial, you will learn how to pre-process a conformer database
file for FastROCS TK, allowing for faster database load times with
OEShapeDatabase.Open. Load times could be up to 10x
faster. See the figure below for an eMolecules dataset of 14 million
conformers.
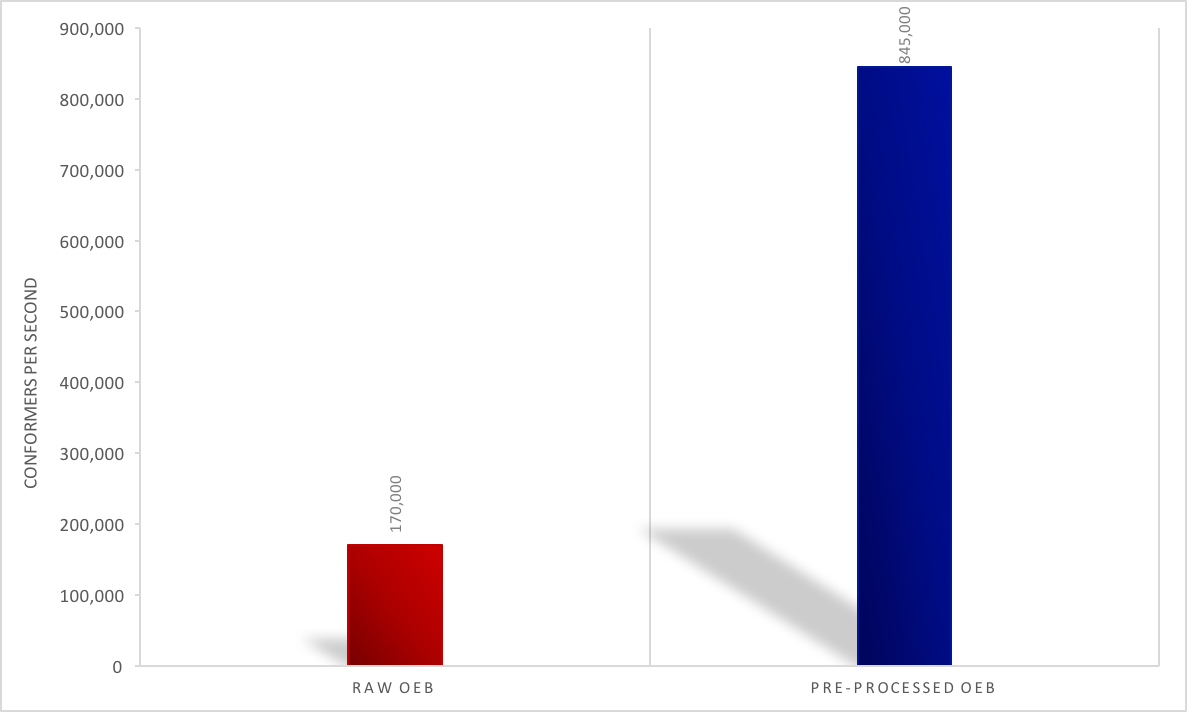
Pre-Processing Performance Impact
To gain this extra loading performance, you need to use the following functions:
OEPreserveRotCompress- this function works on the input molecule stream to ensure that rotor-offset compression is preserved during the preparation process. Rotor offset compression is a way of storing conformers as a set of torsions instead of storing the coordinates for every single conformer of a molecule. This optimization reduces the memory footprint of a multi-conformer molecule.OEPRECompress– this function works on the output molecule stream object allowing the molecules to be stored in a ‘pre-compressed’ format:Writes rotor-offset-compressed molecules in the perfect-rotor-encoding format
There is no need to Gzip which means faster
OEMolDatabase.Open.
OEPrepareFastROCSMol– this function works on each OEMol record of the input.oeb:Sets the energy of each conformer to 0.0 to avoid writing it to OEB.
Suppresses hydrogens and reorders reference conformers for compression.
Pre-calculates color atoms.
Pre-calculates self-color and self-shape terms for all conformers.
Note
The color terms cached by
OEPrepareFastROCSMolare from theOEColorFFType_ImplicitMillsDeancolor force field. A different color force field can be given as the second argument to override ImplicitMillsDean.
In general, calling OEPrepareFastROCSMol and
OEPRECompress will result in a smaller OEB file
than the default OEB.GZ output from OMEGA.
Further reduction in file-size can be achieved by using an
OEMCMolType_HalfFloatCartesian molecule to store
reference coordinates and torsions as 16-bit floating point.
Here is some example code showing how to pre-process a database with
OEPrepareFastROCSMol, save to a precompressed
format, and reduce the file size by using half precision:
For added convenience, we have created a SimplePrepScript.py example script which can be modified to meet your exact needs:
Download code