Matched Pair analysis using a multi-process generation of an MMP index
A program that performs a multi-process matched pair analysis of a set of structures for indexing and saves the generated index file for subsequent loading and querying. This example uses the python multiprocessing module.
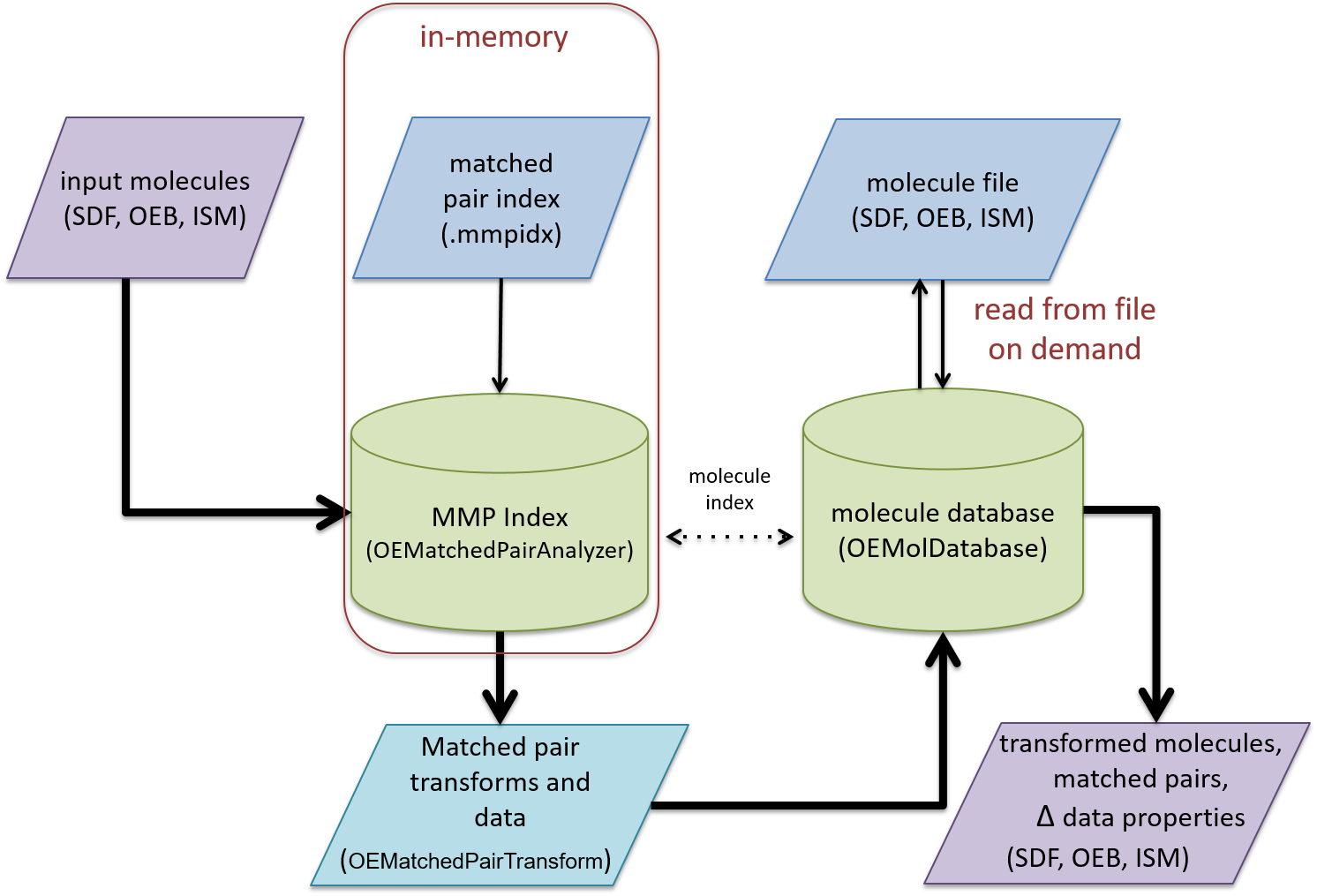
Schematic representation of the Matched Pair Analysis process
See also
OEMatchedPairAnalyzer class
OEMatchedPairApplyTransformsfunction
Command Line Interface
prompt> CreateMMPIndexParallel.py [ -pool num_processes ] index.sdf output.mmpidx
Code
Download code
#!/usr/bin/env python
# (C) 2022 Cadence Design Systems, Inc. (Cadence)
# All rights reserved.
# TERMS FOR USE OF SAMPLE CODE The software below ("Sample Code") is
# provided to current licensees or subscribers of Cadence products or
# SaaS offerings (each a "Customer").
# Customer is hereby permitted to use, copy, and modify the Sample Code,
# subject to these terms. Cadence claims no rights to Customer's
# modifications. Modification of Sample Code is at Customer's sole and
# exclusive risk. Sample Code may require Customer to have a then
# current license or subscription to the applicable Cadence offering.
# THE SAMPLE CODE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND,
# EXPRESS OR IMPLIED. OPENEYE DISCLAIMS ALL WARRANTIES, INCLUDING, BUT
# NOT LIMITED TO, WARRANTIES OF MERCHANTABILITY, FITNESS FOR A
# PARTICULAR PURPOSE AND NONINFRINGEMENT. In no event shall Cadence be
# liable for any damages or liability in connection with the Sample Code
# or its use.
#############################################################################
# Utility to perform a matched pair analysis on a set of structures
# and save the index for subsequent analysis using python multiprocessing
# ---------------------------------------------------------------------------
# CreateMMPIndexParallel.py index_mols output_index
#
# index_mols: filename of input molecules to analyze
# output_index: filename of MMP index
#############################################################################
import sys
import os
import random
import multiprocessing
from tempfile import NamedTemporaryFile
from itertools import islice, chain, repeat
from openeye import oechem
from openeye import oemedchem
BADIDS = []
# globals for slave processes
MMP_INDEX_OPTIONS = 'parallel_mmp_indexing.mmpidx'
OPTIONS_FILE = 'parallel_mmp_options.txt'
def chunk_pad(it, size, padval=None):
it = chain(iter(it), repeat(padval))
return iter(lambda: tuple(islice(it, size)), (padval,) * size)
# MMP index saving is memory intensive - (optional) lock around index serialization
# global definition of write_lock
write_lock = None
def global_write_lock(lck):
global write_lock
write_lock = lck
class CustomOEErrorHandler(oechem.OEErrorHandlerImplBase):
def __init__(self, nowarning):
oechem.OEErrorHandlerImplBase.__init__(self)
self.log = str()
self.nowarning = nowarning
def Msg(self, level, msg):
if level == oechem.OEErrorLevel_Error or level == oechem.OEErrorLevel_Fatal:
self.log += "Preventing call to exit: {0}\n".format(msg)
print("{}: {}".format(oechem.OEErrorLevelToString(level), msg))
sys.exit(1)
elif level == oechem.OEErrorLevel_Verbose:
self.log += "Verbose: {0}\n".format(msg)
elif level == oechem.OEErrorLevel_Warning:
if not self.nowarning:
self.log += "Warning: {0}\n".format(msg)
else:
# Info records are not retained
print("{}: {}".format(oechem.OEErrorLevelToString(level), msg))
def GetLog(self):
return self.log
def CreateCopy(self):
return CustomOEErrorHandler().__disown__()
class FilterSDData:
def __init__(self, fields, asFloating):
if len(fields) == 1 and '-ALLSD' in fields[0].upper():
self.fields = None
elif len(fields) == 1 and '-CLEARSD' in fields[0].upper():
self.fields = []
else:
self.fields = fields
self.asFloating = asFloating
def FilterMolData(self, mol):
if not oechem.OEHasSDData(mol):
return 0
if self.fields is None:
return -1
if len(self.fields) == 0:
oechem.OEClearSDData(mol)
return 0
validdata = 0
deletefields = []
for dp in oechem.OEGetSDDataPairs(mol):
tag = dp.GetTag()
if tag.upper() not in self.fields:
deletefields.append(tag)
continue
value = oechem.OEGetSDData(mol, tag)
if self.asFloating:
try:
float(value)
except ValueError:
oechem.OEThrow.Warning("Failed to convert {} to numeric value ({}) in {}"
.format(tag, value, mol.GetTitle()))
deletefields.append(tag)
continue
validdata += 1
if not validdata:
oechem.OEClearSDData(mol)
else:
for nuke in deletefields:
oechem.OEDeleteSDData(mol, nuke)
return validdata
def mmp_index_mols(args):
idx_opts_fname = args[0]
opts_fname = args[1]
mol_db_fname = args[2]
check_pt_recs = args[3]
idx_opts = oemedchem.OEMatchedPairAnalyzer()
if not oemedchem.OEReadMatchedPairAnalyzer(idx_opts_fname, idx_opts):
oechem.OEThrow.Error("Unable to read index file for options: {0}".format(idx_opts))
return False
keepSD = []
allSD = 1 # default
clearSD = 0
statusrec = 0
stripstereo = 0
stripsalts = 0
dataasfloating = 1 # default
slave_verbose = 0
slave_vverbose = 0
nowarnings = 0
with open(opts_fname, "rt") as OPTS:
for line in OPTS:
line = line.strip().upper()
vals = line.split('=')
if 'KEEPFIELDS' in vals[0]:
keepSD = vals[1].split(',')
allSD = False
clearSD = False
elif 'STATUS' in vals[0]:
statusrec = int(vals[1])
elif 'STEREO' in vals[0]:
stripstereo = int(vals[1])
elif 'SALTS' in vals[0]:
stripsalts = int(vals[1])
elif 'ALLDATA' in vals[0]:
allSD = int(vals[1])
clearSD = not allSD
elif 'ASFLOATING' in vals[0]:
dataasfloating = int(vals[1])
elif 'VERBOSE' in vals[0]:
slave_verbose = int(vals[1])
elif 'VVERBOSE' in vals[0]:
slave_vverbose = int(vals[1])
if slave_vverbose:
slave_verbose = slave_vverbose
elif 'NOWARNINGS' in vals[0]:
nowarnings = int(vals[1])
handler = CustomOEErrorHandler(nowarnings)
owned = False
oechem.OEThrow.SetHandlerImpl(handler, owned)
if allSD:
keepSD = ['-ALLSD']
if clearSD:
keepSD = ['-CLEARSD']
mol = oechem.OEGraphMol()
moldb = oechem.OEMolDatabase()
if not moldb.Open(mol_db_fname):
oechem.OEThrow.Error("Unable to open molecule database: {0}".format(mol_db_fname))
return False
# interpret floating point data if requested
validdata = FilterSDData(keepSD, dataasfloating)
# CREATE the index using the passed options template
mmpidx = oemedchem.OEMatchedPairAnalyzer(idx_opts.GetOptions())
num_mols = 0
CHECK_POINT_FILE = None
if check_pt_recs > 0:
CHECK_POINT_FILE = NamedTemporaryFile(prefix="checkpoint_", suffix='.mmpidx', delete=False)
mmpidxEXT = '.mmpidx'
# GENERATE a tempfile name to return the serialized index
with NamedTemporaryFile(prefix="slave_", suffix=mmpidxEXT, delete=False) as MMP_OUT_IDX:
unindexed = 0
timer = oechem.OEStopwatch()
numrec = 0
for molid in args[4:]:
# end of list may be padded with None
if molid is None:
break
numrec += 1
idx = int(molid)
if not moldb.GetMolecule(mol, idx):
oechem.OEThrow.Warning('Error retrieving record={0}'.format(idx))
continue
if mol.GetTitle() and mol.GetTitle in BADIDS:
oechem.OEThrow.Warning('{}: skipping bad structure: {}'.format(idx, mol.GetTitle()))
continue
if not allSD:
# filter the input molecule based on "keeper" SD data fields
validdata.FilterMolData(mol)
if stripsalts:
oechem.OEDeleteEverythingExceptTheFirstLargestComponent(mol)
if stripstereo:
oechem.OEUncolorMol(mol,
(oechem.OEUncolorStrategy_RemoveAtomStereo |
oechem.OEUncolorStrategy_RemoveBondStereo))
# add molecule to the index
status = mmpidx.AddMol(mol, idx)
if status == idx:
num_mols += 1
else:
oechem.OEThrow.Warning('Error indexing molecule {}: {}: {}'
.format(idx, oemedchem.OEMatchedPairIndexStatusName(status),
mol.GetTitle()))
unindexed += 1
if statusrec > 0 and (numrec % statusrec) == 0:
if slave_verbose:
oechem.OEThrow.Info('Records: {}, Indexed: {}, Unindexed: {}, Indexing rate: {:.1F} mol/sec'
.format(numrec, num_mols, unindexed,
float(numrec) / timer.Elapsed()))
else:
oechem.OEThrow.Info('Records: {}, Indexed: {}, Unindexed: {}'
.format(numrec, num_mols, unindexed))
if check_pt_recs > 0 and (numrec % check_pt_recs) == 0:
oechem.OEThrow.Info('Records: {}, Checkpointing: {}'
.format(numrec, CHECK_POINT_FILE.name))
if write_lock is not None:
write_lock.acquire()
bcheckpt = oemedchem.OEWriteMatchedPairAnalyzer(CHECK_POINT_FILE.name, mmpidx)
if write_lock is not None:
write_lock.release()
oechem.OEThrow.Info('Records: {}, Checkpointed: {}'
.format(numrec, CHECK_POINT_FILE.name))
if CHECK_POINT_FILE is not None:
CHECK_POINT_FILE.close()
if slave_vverbose:
oechem.OEThrow.Info('Records: {}, serialization in progress...{}'
.format(numrec, MMP_OUT_IDX.name))
bcheckpt = oemedchem.OEWriteMatchedPairAnalyzer(MMP_OUT_IDX.name, mmpidx)
if slave_vverbose:
oechem.OEThrow.Info('Records: {}, serialization complete.'
.format(numrec))
if bcheckpt:
if CHECK_POINT_FILE is not None:
os.remove(CHECK_POINT_FILE.name)
else:
return (False, None, unindexed, handler.GetLog())
return (True, MMP_OUT_IDX.name, unindexed, handler.GetLog())
def MMPIndex(itf):
# input structures to index
MOL_DB_IDX = itf.GetString("-input")
verbose = itf.GetBool("-verbose")
vverbose = itf.GetBool("-vverbose")
if vverbose:
verbose = True
# create the moldatabase index on the input structures so the slaves can access it
if not oechem.OECreateMolDatabaseIdx(MOL_DB_IDX):
oechem.OEThrow.Fatal("Unable to generate molecule database for {}"
.format(itf.GetString("-input")))
# output index file
mmpindexfile = itf.GetString("-output")
if not oemedchem.OEIsMatchedPairAnalyzerFileType(mmpindexfile):
oechem.OEThrow.Fatal("Output file is not a matched pair index type - needs \
.mmpidx extension: {}"
.format(mmpindexfile))
moldb = oechem.OEMolDatabase()
if not moldb.Open(MOL_DB_IDX):
oechem.OEThrow.Fatal("Unable to open molecule database: {}"
.format(MOL_DB_IDX))
# create options class with defaults
opts = oemedchem.OEMatchedPairAnalyzerOptions()
# setup options from command line
if not oemedchem.OESetupMatchedPairIndexOptions(opts, itf):
oechem.OEThrow.Fatal("Error setting matched pair indexing options!")
if verbose:
if not opts.HasIndexableFragmentHeavyAtomRange():
oechem.OEThrow.Info("Indexing all fragments")
else:
oechem.OEThrow.Info("Limiting fragment cores to {0:.2f}-{1:.2f}% of input molecules"
.format(opts.GetIndexableFragmentRangeMin(),
opts.GetIndexableFragmentRangeMax()))
if (opts.GetOptions() &
oemedchem.OEMatchedPairOptions_UniquesOnly) != 0 and itf.GetInt("-pool") != 1:
if not itf.GetBool("-uniqueinput"):
oechem.OEThrow.Warning("Disabling uniqueness check for multi-process indexing, use -uniqueinput instead")
if not opts.SetOptions(opts.GetOptions() & ~oemedchem.OEMatchedPairOptions_UniquesOnly):
oechem.OEThrow.Fatal("Error disabling indexer -unique option")
# create indexing engine
mmpidx = oemedchem.OEMatchedPairAnalyzer(opts)
# dump the index so the slaves can load the options from it
if not oemedchem.OEWriteMatchedPairAnalyzer(MMP_INDEX_OPTIONS, mmpidx):
oechem.OEThrow.Fatal("Unable to serialize index for %s" % MMP_INDEX_OPTIONS)
maxrecOPT = max(itf.GetInt("-maxrec"), 0)
checkptrec = itf.GetInt("-checkpoint")
statusrec = itf.GetInt("-status")
stripstereo = itf.GetBool("-stripstereo")
if stripstereo and verbose:
oechem.OEThrow.Info("Stripping stereo")
stripsalts = itf.GetBool("-stripsalts")
if stripsalts and verbose:
oechem.OEThrow.Info("Stripping salts")
keepFields = []
if itf.HasString("-keepSD"):
for field in itf.GetStringList("-keepSD"):
keepFields.append(field)
if verbose:
oechem.OEThrow.Info('Retaining SD data fields: {}'.format(' '.join(keepFields)))
alldata = itf.GetBool("-allSD")
cleardata = itf.GetBool("-clearSD")
if keepFields:
if verbose and (alldata or cleardata):
oechem.OEThrow.Info("Option -keepSD overriding -allSD, -clearSD")
alldata = False
cleardata = False
elif cleardata:
alldata = False
if verbose:
oechem.OEThrow.Info("Forced clearing of all input SD data")
else:
if verbose:
if not alldata:
oechem.OEThrow.Info("No SD data handling option specified, -allSD assumed")
else:
oechem.OEThrow.Info("Retaining all input SD data")
alldata = True
cleardata = False
with open(OPTIONS_FILE, "wt") as WRITE_OPTS:
if cleardata:
WRITE_OPTS.write('ALLDATA=0\n')
elif alldata:
WRITE_OPTS.write('ALLDATA=1\n')
elif keepFields:
WRITE_OPTS.write('KEEPFIELDS={0}\n'.format(','.join(keepFields)))
WRITE_OPTS.write('STATUS={0}\n'.format(statusrec))
if stripstereo:
WRITE_OPTS.write('STEREO=1\n')
if stripsalts:
WRITE_OPTS.write('SALTS=1\n')
if verbose:
WRITE_OPTS.write('VERBOSE=1\n')
if vverbose:
WRITE_OPTS.write('VVERBOSE=1\n')
if itf.GetBool("-nowarnings"):
WRITE_OPTS.write('NOWARNINGS=1\n')
# get list of record ids
if not maxrecOPT:
maxrec = moldb.GetMaxMolIdx()
else:
maxrec = maxrecOPT
if not itf.GetBool("-uniqueinput"):
molids = [i for i in range(0, maxrec)]
else:
mol = oechem.OEGraphMol()
umols = set()
molids = []
for i in range(0, maxrec):
if not moldb.GetMolecule(mol, i):
continue
if stripsalts:
oechem.OEDeleteEverythingExceptTheFirstLargestComponent(mol)
if stripstereo:
oechem.OEUncolorMol(mol,
(oechem.OEUncolorStrategy_RemoveAtomStereo |
oechem.OEUncolorStrategy_RemoveBondStereo))
smi = oechem.OEMolToSmiles(mol)
if smi in umols:
continue
umols.add(smi)
molids.append(i)
if maxrecOPT and verbose:
oechem.OEThrow.Info("Indexing a maximum of {} records"
.format(len(molids)))
nrprocesses = itf.GetInt("-pool")
if not nrprocesses:
nrprocesses = oechem.OEGetNumProcessors()
# identify warning handling, etc
if verbose:
if itf.GetBool("-exportcompress"):
oechem.OEThrow.Info("Removing singleton index nodes from index")
if not itf.GetInt("-pool"):
oechem.OEThrow.Info("Using the maximum number of processes allowed ({})".format(nrprocesses))
else:
oechem.OEThrow.Info("Limiting indexing to {} process(es)".format(nrprocesses))
if itf.GetBool("-nowarnings"):
oechem.OEThrow.Info("Suppressing warnings during indexing")
else:
oechem.OEThrow.Info("Emitting warnings during indexing")
# randomize indices to avoid pathologicals near each other
if not itf.GetBool("-randomize"):
if verbose:
oechem.OEThrow.Info("Sequentially processing input records for indexing")
else:
if verbose:
oechem.OEThrow.Info("Randomizing input records for indexing")
random.shuffle(molids)
if checkptrec:
oechem.OEThrow.Info("Checkpointing indices after every {0} records"
.format(checkptrec))
if statusrec:
oechem.OEThrow.Info("Status output after every {0} records"
.format(statusrec))
test_input = list(chunk_pad(molids,
int((len(molids) / nrprocesses) + (len(molids) % nrprocesses))))
# prepend the filenames to for loading and extraction index options
test_input = [[MMP_INDEX_OPTIONS, OPTIONS_FILE, MOL_DB_IDX, checkptrec]
+ list(i) for i in test_input]
# begin molecule indexing
timer = oechem.OEStopwatch()
# creating the indexing pool - (optional)
# lock can limit index serialization to ONE process at a time
# mlock = multiprocessing.Lock()
mlock = None
processes = multiprocessing.Pool(nrprocesses, initializer=global_write_lock, initargs=(mlock,))
errs = None
if itf.GetBool("-nowarnings"):
errs = oechem.oeosstream()
oechem.OEThrow.SetOutputStream(errs)
results = []
for i, res in enumerate(processes.imap_unordered(mmp_index_mols, test_input)):
results.append(res)
processes.close()
# restore OEThrow output
oechem.OEThrow.SetOutputStream(oechem.oeout)
# capture indexing time
tIndexing = timer.Elapsed()
mmpidx = oemedchem.OEMatchedPairAnalyzer()
# walk the results and read in the indexed segments for the input molecules
unindexed = 0
for res in results:
(status, idxfile, seg_unindexed, log) = res
if vverbose:
oechem.OEThrow.Info('status: {0} index: {1} unindexed: {2}'
.format(status, idxfile, seg_unindexed))
if verbose and log:
oechem.OEThrow.Info(log)
if not status:
continue
curmols = mmpidx.NumMols()
curmmps = mmpidx.NumMatchedPairs()
mergetimer = oechem.OEStopwatch()
if not oemedchem.OEReadMatchedPairAnalyzer(idxfile, mmpidx, True):
oechem.OEThrow.Warning('Error reading {}'.format(idxfile))
if errs is not None:
errs.clear()
os.remove(idxfile)
if vverbose:
oechem.OEThrow.Info("{0}: {1:.2F} sec added mols: {2:,} added unindexed: {3:,} added mmps: {4:,} "
.format(idxfile, mergetimer.Elapsed(),
mmpidx.NumMols() - curmols,
seg_unindexed,
mmpidx.NumMatchedPairs() - curmmps))
unindexed += seg_unindexed
# capture index merge time
tMerging = timer.Elapsed() - tIndexing
if not mmpidx.NumMols():
oechem.OEThrow.Fatal('No records in index structure file')
if not mmpidx.NumMatchedPairs() or unindexed == maxrec:
oechem.OEThrow.Fatal('No matched pairs found from indexing, ' +
'use -fragGe,-fragLe options to extend index range')
numrec = len(molids)
if verbose:
oechem.OEThrow.Info("Indexing time: {0:.2F} indexing rate: {1:.2F} mol/sec"
.format(tIndexing, float(numrec) / tIndexing))
oechem.OEThrow.Info("Index merge time: {0:.2F} indexing merge rate: {1:.2F} mol/sec"
.format(tMerging, float(numrec) / tMerging))
oechem.OEThrow.Info("Total time: {0:.2F} total rate: {1:.1F} mol/sec"
.format(timer.Elapsed(), float(numrec) / timer.Elapsed()))
# return some status information
oechem.OEThrow.Info("Records: {}, Indexed: {}, matched pairs: {:,d}"
.format(numrec,
mmpidx.NumMols(),
mmpidx.NumMatchedPairs()))
if itf.GetBool("-exportcompress"):
if not mmpidx.ModifyOptions(oemedchem.OEMatchedPairOptions_ExportCompression, 0):
oechem.OEThrow.Warning("Error enabling export compression!")
# export merged index
if not oemedchem.OEWriteMatchedPairAnalyzer(mmpindexfile, mmpidx):
oechem.OEThrow.Fatal('Error serializing MMP index: {}'.format(mmpindexfile))
# cleanup
try:
os.remove(MMP_INDEX_OPTIONS)
except OSError:
pass
try:
os.remove(OPTIONS_FILE)
except OSError:
pass
# remove the moldatabase index on the input structures
try:
os.remove(oechem.OEGetMolDatabaseIdxFileName(MOL_DB_IDX))
except OSError:
pass
return 0
############################################################
InterfaceData = """
# createmmpindexparallel interface file
!CATEGORY CreateMMPIndexParallel
!CATEGORY I/O
!PARAMETER -input 1
!ALIAS -in
!TYPE string
!REQUIRED true
!BRIEF Input filename of structures to index
!KEYLESS 1
!END
!PARAMETER -output 2
!ALIAS -out
!TYPE string
!REQUIRED true
!BRIEF Output filename for serialized MMP index
!KEYLESS 2
!END
!END
!CATEGORY indexing_options
!PARAMETER -maxrec 1
!TYPE int
!DEFAULT 0
!LEGAL_RANGE 0 inf
!BRIEF process at most -maxrec records from -input (0: all)
!END
!PARAMETER -status 2
!TYPE int
!DEFAULT 0
!LEGAL_RANGE 0 inf
!BRIEF emit progress information every -status records (0: off)
!END
!PARAMETER -pool 3
!TYPE int
!DEFAULT 0
!LEGAL_RANGE 0 inf
!BRIEF sets the number of parallel workers, \
(0: use value returned from OEGetNumProcessors())
!END
!PARAMETER -uniqueinput 4
!TYPE bool
!DEFAULT 0
!BRIEF discard duplicate input structures after -stripsalts, -stripstereo activities
!END
!PARAMETER -exportcompress 5
!TYPE bool
!DEFAULT 0
!BRIEF Whether to remove singleton nodes on export of the MMP index
!DETAIL
True indicates no additional structures will be added to the index
!END
!PARAMETER -randomize 6
!TYPE bool
!DEFAULT 0
!BRIEF randomize input records
!END
!PARAMETER -checkpoint 7
!TYPE int
!LEGAL_RANGE 0 inf
!DEFAULT 0
!BRIEF checkpoint the index segments every -checkpoint records
!END
!PARAMETER -verbose 8
!TYPE bool
!DEFAULT 0
!BRIEF generate verbose output
!END
!PARAMETER -vverbose 9
!TYPE bool
!DEFAULT 0
!BRIEF generate very verbose output
!END
!END
!CATEGORY molecule_SDData
!PARAMETER -allSD 1
!TYPE bool
!DEFAULT 0
!BRIEF retain all input SD data
!END
!PARAMETER -clearSD 2
!TYPE bool
!DEFAULT 0
!BRIEF clear all input SD data
!END
!PARAMETER -keepSD 3
!TYPE string
!LIST true
!BRIEF list of SD data tags of floating point data to \
*retain* for indexing (all other SD data is removed)
!END
!END
!CATEGORY molecule_processing
!PARAMETER -stripstereo 1
!TYPE bool
!DEFAULT 0
!BRIEF Whether to strip stereo from -input structures
!END
!PARAMETER -stripsalts 2
!TYPE bool
!DEFAULT 0
!BRIEF Whether to strip salt fragments from -input structures
!END
!PARAMETER -nowarnings 3
!TYPE bool
!DEFAULT 1
!BRIEF suppress warning messages from reading -input (default: True)
!END
!END
!END
"""
def main(argv=[__name__]):
itf = oechem.OEInterface(InterfaceData)
oemedchem.OEConfigureMatchedPairIndexOptions(itf)
if not oechem.OEParseCommandLine(itf, argv):
oechem.OEThrow.Fatal("Unable to interpret command line!")
MMPIndex(itf)
if __name__ == "__main__":
sys.exit(main(sys.argv))