Matched Pair analysis using a multi-threaded generation of a MMP index
A program that performs a multi-threaded matched pair analysis of a set of structures for indexing and saves the generated index file for subsequent loading and querying.
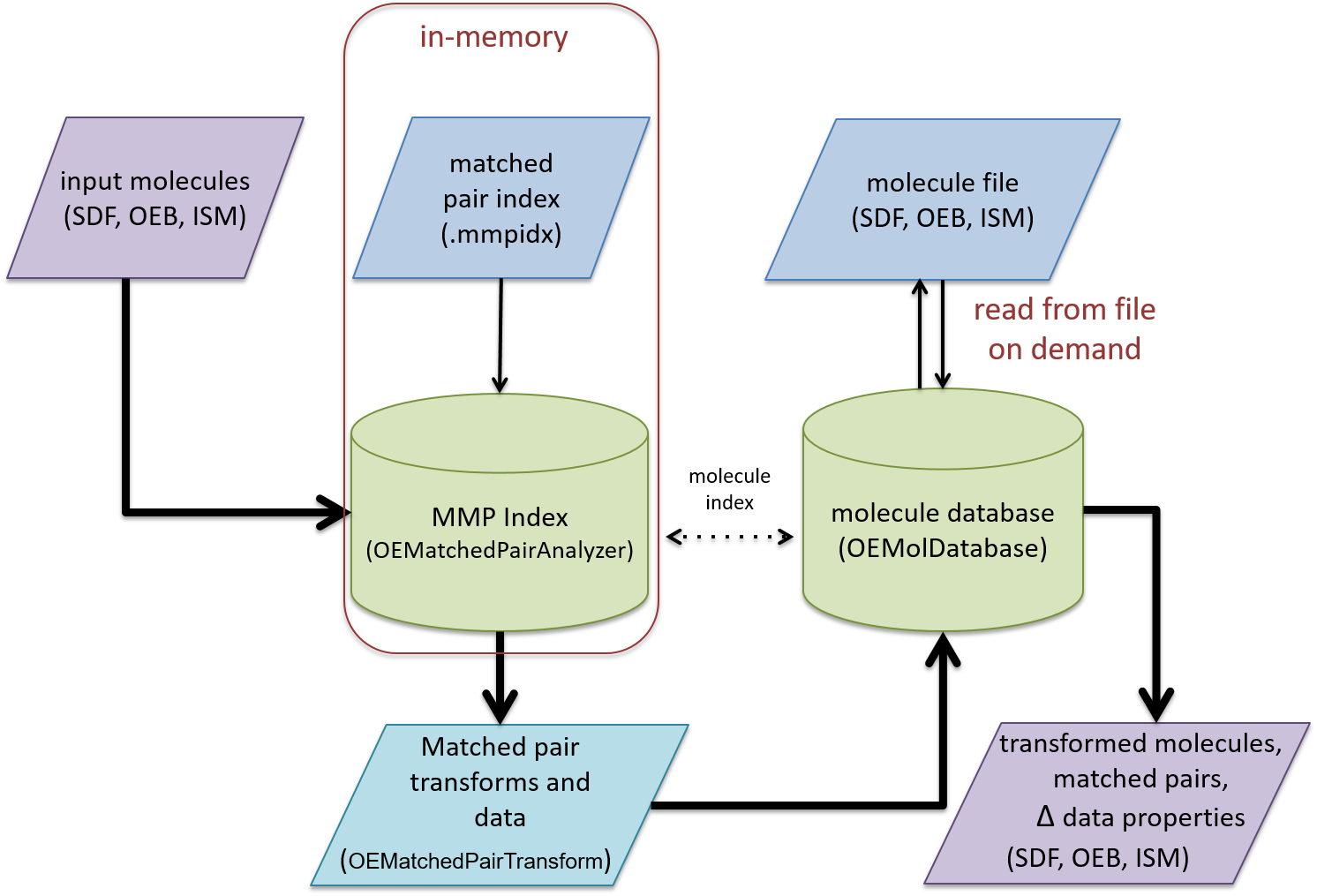
Schematic representation of the Matched Pair Analysis process
See also
OEMatchedPairAnalyzer class
OEMatchedPairApplyTransformsfunction
Command Line Interface
prompt> CreateMMPIndexThreaded.py [ -threads num_threads ] index.sdf output.mmpidx
Code
Download code
#!/usr/bin/env python
# (C) 2022 Cadence Design Systems, Inc. (Cadence)
# All rights reserved.
# TERMS FOR USE OF SAMPLE CODE The software below ("Sample Code") is
# provided to current licensees or subscribers of Cadence products or
# SaaS offerings (each a "Customer").
# Customer is hereby permitted to use, copy, and modify the Sample Code,
# subject to these terms. Cadence claims no rights to Customer's
# modifications. Modification of Sample Code is at Customer's sole and
# exclusive risk. Sample Code may require Customer to have a then
# current license or subscription to the applicable Cadence offering.
# THE SAMPLE CODE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND,
# EXPRESS OR IMPLIED. OPENEYE DISCLAIMS ALL WARRANTIES, INCLUDING, BUT
# NOT LIMITED TO, WARRANTIES OF MERCHANTABILITY, FITNESS FOR A
# PARTICULAR PURPOSE AND NONINFRINGEMENT. In no event shall Cadence be
# liable for any damages or liability in connection with the Sample Code
# or its use.
#############################################################################
# Utility to perform a matched pair analysis on a set of structures
# and save the index for subsequent analysis using a multithreaded API
# ---------------------------------------------------------------------------
# CreateMMPIndexThreaded.py index_mols output_index
#
# index_mols: filename of input molecules to analyze
# output_index: filename of MMP index
#############################################################################
from openeye import oechem
from openeye import oemedchem
import sys
def MMPIndex(itf):
# checking input structures
ifsindex = oechem.oemolistream()
if not ifsindex.open(itf.GetString("-input")):
oechem.OEThrow.Fatal("Unable to open {} for reading"
.format(itf.GetString("-input")))
ifsindex.close()
verbose = itf.GetBool("-verbose")
vverbose = itf.GetBool("-vverbose")
if vverbose:
verbose = True
# output index file
mmpindexfile = itf.GetString("-output")
if not oemedchem.OEIsMatchedPairAnalyzerFileType(mmpindexfile):
oechem.OEThrow.Fatal("Output file is not a matched pair index type - \
needs .mmpidx extension: {}" .format(mmpindexfile))
# create options class with defaults
mmpopts = oemedchem.OEMatchedPairAnalyzerOptions()
# set up options from command line
if not oemedchem.OESetupMatchedPairIndexOptions(mmpopts, itf):
oechem.OEThrow.Fatal("Error setting matched pair indexing options!")
if verbose:
if not mmpopts.HasIndexableFragmentHeavyAtomRange():
oechem.OEThrow.Info("Indexing all fragments")
else:
oechem.OEThrow.Info("Limiting fragment cores to {0:.2f}-{1:.2f}% of input molecules"
.format(mmpopts.GetIndexableFragmentRangeMin(),
mmpopts.GetIndexableFragmentRangeMax()))
if itf.GetInt("-maxrec") and verbose:
oechem.OEThrow.Info("Indexing a maximum of {} records"
.format(itf.GetInt("-maxrec")))
if itf.GetBool("-exportcompress"):
if verbose:
oechem.OEThrow.Info("Removing singleton index nodes from index")
if not mmpopts.SetOptions(mmpopts.GetOptions() |
oemedchem.OEMatchedPairOptions_ExportCompression):
oechem.OEThrow.Warning("Error enabling export compression!")
# set indexing options
indexopts = oemedchem.OECreateMMPIndexOptions(mmpopts)
# set requested verbosity setting
if vverbose:
indexopts.SetVerbose(2)
elif verbose:
indexopts.SetVerbose(1)
# limit number of records to process
indexopts.SetMaxRecord(itf.GetInt("-maxrec"))
# set number of threads to use
indexopts.SetNumThreads(itf.GetInt("-threads"))
if verbose:
if not indexopts.GetNumThreads():
oechem.OEThrow.Info("Using the maximum number of threads available")
else:
oechem.OEThrow.Info("Limiting indexing to {} thread(s)"
.format(indexopts.GetNumThreads()))
errs = None
if itf.GetBool("-nowarnings"):
errs = oechem.oeosstream()
oechem.OEThrow.SetOutputStream(errs)
if verbose:
oechem.OEThrow.Info("Threaded indexing of {}, all SD data will be preserved".format(itf.GetString("-input")))
# create index
indexstatus = oemedchem.OECreateMMPIndexFile(mmpindexfile,
itf.GetString("-input"),
indexopts)
dupes = 0
if errs is not None:
oechem.OEThrow.SetOutputStream(oechem.oeout)
for err in errs.str().decode().split('\n'):
err = err.rstrip()
if not err:
continue
if verbose:
oechem.OEThrow.Info(err)
if 'ignoring duplicate molecule,' in err:
dupes += 1
if not indexstatus.IsValid():
oechem.OEThrow.Fatal('Invalid status returned from indexing!')
if not indexstatus.GetTotalMols():
oechem.OEThrow.Fatal('No records in index structure file: {}'
.format(itf.GetString("-input")))
if dupes:
oechem.OEThrow.Info('Found {} duplicate structures during indexing'
.format(dupes))
if not indexstatus.GetNumMatchedPairs():
oechem.OEThrow.Fatal('No matched pairs found from indexing, ' +
'use -fragGe,-fragLe options to extend indexing range')
# return some status information
oechem.OEThrow.Info("Records: {}, Indexed: {}, matched pairs: {:,d}"
.format(indexstatus.GetTotalMols(),
indexstatus.GetNumMols(),
indexstatus.GetNumMatchedPairs()))
return 0
############################################################
InterfaceData = """
# createmmpindexthreaded interface file
!CATEGORY CreateMMPIndexThreaded
!CATEGORY I/O
!PARAMETER -input 1
!ALIAS -in
!TYPE string
!REQUIRED true
!BRIEF Input filename of structures to index
!KEYLESS 1
!END
!PARAMETER -output 2
!ALIAS -out
!TYPE string
!REQUIRED true
!BRIEF Output filename for serialized MMP index
!KEYLESS 2
!END
!END
!CATEGORY indexing_options
!PARAMETER -maxrec 1
!TYPE int
!DEFAULT 0
!LEGAL_RANGE 0 inf
!BRIEF process at most -maxrec records from -input (0: all)
!END
!PARAMETER -threads 2
!TYPE int
!DEFAULT 0
!LEGAL_RANGE 0 inf
!BRIEF limit number of indexing threads to -threads (0:default)
!END
!PARAMETER -exportcompress 3
!TYPE bool
!DEFAULT 0
!BRIEF Whether to remove singleton nodes on export of the MMP index
!DETAIL
True indicates no additional structures will be added to the index
!END
!PARAMETER -nowarnings 4
!TYPE bool
!DEFAULT 1
!BRIEF suppress warning messages from indexing -input (default: True)
!END
!PARAMETER -verbose 5
!TYPE bool
!DEFAULT 0
!BRIEF generate verbose output
!END
!PARAMETER -vverbose 6
!TYPE bool
!DEFAULT 0
!BRIEF generate very verbose output
!END
!END
!END
"""
def main(argv=[__name__]):
itf = oechem.OEInterface(InterfaceData)
oemedchem.OEConfigureMatchedPairIndexOptions(itf)
if not oechem.OEParseCommandLine(itf, argv):
oechem.OEThrow.Fatal("Unable to interpret command line!")
MMPIndex(itf)
if __name__ == "__main__":
sys.exit(main(sys.argv))