MCS Similarity score reporting
A program that loads a previously generated MCS fragment database, then returns the highest Maximum Common Substructure (Tanimoto) scores from the provided input query structures.
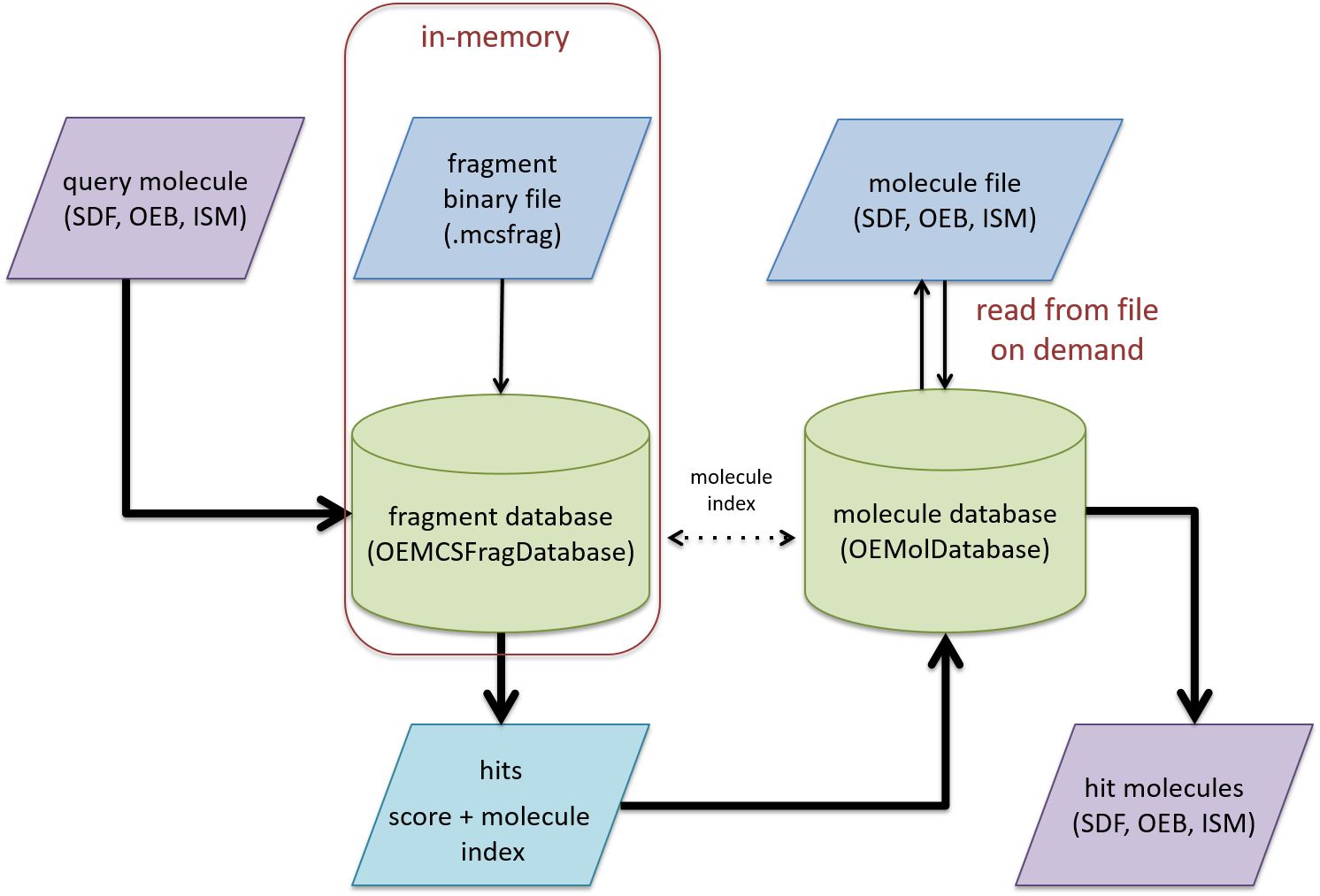
Schematic representation of the MCS fragment index and search process
See also
OEMCSFragDatabase class
OEMCSFragDatabaseOptions class
Command Line Interface
prompt> MCSFragDatabase.py -indb index.mcsfrag -query queries.sdf
Code
Download code
#!/usr/bin/env python
# (C) 2022 Cadence Design Systems, Inc. (Cadence)
# All rights reserved.
# TERMS FOR USE OF SAMPLE CODE The software below ("Sample Code") is
# provided to current licensees or subscribers of Cadence products or
# SaaS offerings (each a "Customer").
# Customer is hereby permitted to use, copy, and modify the Sample Code,
# subject to these terms. Cadence claims no rights to Customer's
# modifications. Modification of Sample Code is at Customer's sole and
# exclusive risk. Sample Code may require Customer to have a then
# current license or subscription to the applicable Cadence offering.
# THE SAMPLE CODE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND,
# EXPRESS OR IMPLIED. OPENEYE DISCLAIMS ALL WARRANTIES, INCLUDING, BUT
# NOT LIMITED TO, WARRANTIES OF MERCHANTABILITY, FITNESS FOR A
# PARTICULAR PURPOSE AND NONINFRINGEMENT. In no event shall Cadence be
# liable for any damages or liability in connection with the Sample Code
# or its use.
###############################################################################
# Utility to load a pregenerated index and return the highest MCS similarity
# structures from an query structure(s)
# ---------------------------------------------------------------------------
# MCSFragDatabase [ -indb index.mcsfrag ]
# [ -query query_file ]
# [ -type bond -limit 10 ] // 10 Tanimoto sim bond scores
# [ -verbose 1 ] // optional verbosity
#
# index.mcsfrag: filename of index file to load
# query_file: filename of query to report MCS similarity results
###############################################################################
from openeye import oechem
from openeye import oemedchem
import sys
def MCSFragQuery(itf):
verbose = itf.GetBool("-verbose")
timer = itf.GetBool("-timer")
indb = itf.GetString("-indb")
if not oemedchem.OEIsMCSFragDatabaseFileType(indb):
oechem.OEThrow.Fatal("Option -indb must specify a .mcsfrag filename: "
+ indb)
if verbose:
oechem.OEThrow.Info("Loading index from " + indb)
watch = oechem.OEStopwatch()
watch.Start()
mcsdb = oemedchem.OEMCSFragDatabase()
if not oemedchem.OEReadMCSFragDatabase(indb, mcsdb):
oechem.OEThrow.Fatal("Error deserializing MCS fragment database: "
+ indb)
loadtime = watch.Elapsed()
if not mcsdb.NumMols():
oechem.OEThrow.Fatal("Loaded empty index")
if timer:
if not loadtime:
oechem.OEThrow.Info("Loaded index of {0} molecules, {1} fragments"
.format(mcsdb.NumMols(), mcsdb.NumFragments()))
else:
oechem.OEThrow.Info("Loaded index of {0} molecules, {1} "
"fragments in {2:.2F} sec: "
"{3:,.1F} mols/sec {4:,.1F} frags/sec"
.format(mcsdb.NumMols(),
mcsdb.NumFragments(),
loadtime,
float(mcsdb.NumMols())/loadtime,
float(mcsdb.NumFragments())/loadtime))
if not mcsdb.NumFragments():
oechem.OEThrow.Fatal("No fragments loaded from index, "
"use -fragGe,-fragLe options to "
"extend indexable range")
queryfile = itf.GetString("-query")
ifsquery = oechem.oemolistream()
if not ifsquery.open(queryfile):
oechem.OEThrow.Fatal("Unable to open query file: " + queryfile)
# process the query options
qmaxrec = 0
if itf.HasInt("-qlim"):
qmaxrec = itf.GetInt("-qlim")
numscores = 10
if itf.HasInt("-limit"):
numscores = itf.GetInt("-limit")
cnttype = oemedchem.OEMCSScoreType_BondCount
if itf.HasString("-type"):
if "atom" in itf.GetString("-type"):
cnttype = oemedchem.OEMCSScoreType_AtomCount
elif "bond" in itf.GetString("-type"):
cnttype = oemedchem.OEMCSScoreType_BondCount
else:
oechem.OEThrow.Warning("Ignoring unrecognized -type option, "
"expecting atom or bond: " +
itf.GetString("-type"))
cntname = "bond"
if cnttype != oemedchem.OEMCSScoreType_BondCount:
cntname = "atom"
# process the query (or queries)
qmol = oechem.OEGraphMol()
qnum = 0
while oechem.OEReadMolecule(ifsquery, qmol):
qnum += 1
numhits = 0
for score in mcsdb.GetSortedScores(qmol,
numscores,
0,
0,
True,
cnttype):
if numhits == 0:
oechem.OEThrow.Info("Query: {0}: {1}"
.format(qnum, qmol.GetTitle()))
oechem.OEThrow.Info("\trecord: {0:6}\ttanimoto_{1}_score: "
"{2:.2f}\tmcs_core: {3}"
.format(score.GetIdx(),
cntname,
score.GetScore(),
score.GetMCSCore()))
numhits += 1
if numhits == 0:
oechem.OEThrow.Warning("Query: {0}: {1} - no hits"
.format(qnum, qmol.GetTitle()))
if qmaxrec and qnum >= qmaxrec:
break
if qnum == 0:
oechem.OEThrow.Fatal("Error reading query structure(s): " + queryfile)
############################################################
InterfaceData = """
#MCSFragmentDatabase interface file
!CATEGORY MCSFragmentDatabase
!CATEGORY I/O
!PARAMETER -indb 1
!TYPE string
!REQUIRED true
!BRIEF Input filename of index file to load
!KEYLESS 1
!END
!PARAMETER -query 2
!ALIAS -q
!TYPE string
!REQUIRED true
!BRIEF query structure file to report similarity results against
!KEYLESS 2
!END
!END
!CATEGORY options
!PARAMETER -type 1
!TYPE string
!DEFAULT bond
!BRIEF MCS core counts to use for reported scores: atom or bond
!END
!PARAMETER -limit 2
!TYPE int
!DEFAULT 10
!BRIEF report -limit scores for the query structure
!END
!PARAMETER -verbose 3
!TYPE bool
!DEFAULT 0
!BRIEF generate verbose output
!END
!PARAMETER -qlim 4
!TYPE int
!DEFAULT 0
!BRIEF limit query processing to -qlim records from the -query structures
!END
!PARAMETER -timer 5
!TYPE bool
!DEFAULT 0
!BRIEF report database loading time
!END
!END
!END
"""
def main(argv=[__name__]):
itf = oechem.OEInterface(InterfaceData)
if not oechem.OEParseCommandLine(itf, argv):
oechem.OEThrow.Fatal("Unable to interpret command line!")
MCSFragQuery(itf)
if __name__ == "__main__":
sys.exit(main(sys.argv))