Valence Models
This section describes the three valence models currently implemented by OEChem TK.
For molecules that have fully specified formal charges, the MDL valence model may be used to assign hydrogen counts. (See section MDL Valence Model.)
For molecules that have fully specified hydrogen counts, the OpenEye ‘’charge’’ model may be used to assign formal charges. (See section OpenEye Charge Model.)
Finally, for molecules with neither formal charges nor hydrogen counts, OEChem TK uses the OpenEye hydrogen count model to assign both hydrogen counts and formal charges. (See section OpenEye Hydrogen Count Model.)
MDL Valence Model
The MDL valence model was developed by MDL to allow hydrogen counts to be implicit in MDL SD and MOL file formats. MDL valence is simply the number of covalent bonds to an atom (i.e., the sum of the bond orders), not the number of bonded neighbors. It assumes that the bond orders to an atom are specified (explicit valence), and that the atomic number and formal charge are correctly set. The MDL valence model then prescribes the number of implicit hydrogens on a particular atom. The periodic table graphic below shows the MDL valence model as implemented in OEChem TK.
The valence rules are,
For transition metals, lanthanides and actinides, all valences are allowed which means that implicit hydrogen counts will never be applied.
For the main group elements, allowed valences are listed parenthetically for each atom in the graphic below.
Charges are handled by a simple shift: for positive and negative charges, convert the charged atom to its isoelectronic neutral atom; positive charges move atoms to the left and negative charges move the atom to the right in the periodic table, skipping the transition metal block. Thus,
C+is considered to be isoelectronic toB,C(+2)is equivalent toBe,C(-1)is equivalent toN, andC(-2)is equivalent toO.If the isolectronic version of a charged atom is a Group I or II atom, no implicit hydrogen counts are applied (e.g.,
Sn(+2))A charged atom is considered an illegal valence if its neutral isolectronic atom type wraps beyond a noble gas configuration (e.g.,
Li(+2)orF(-2))
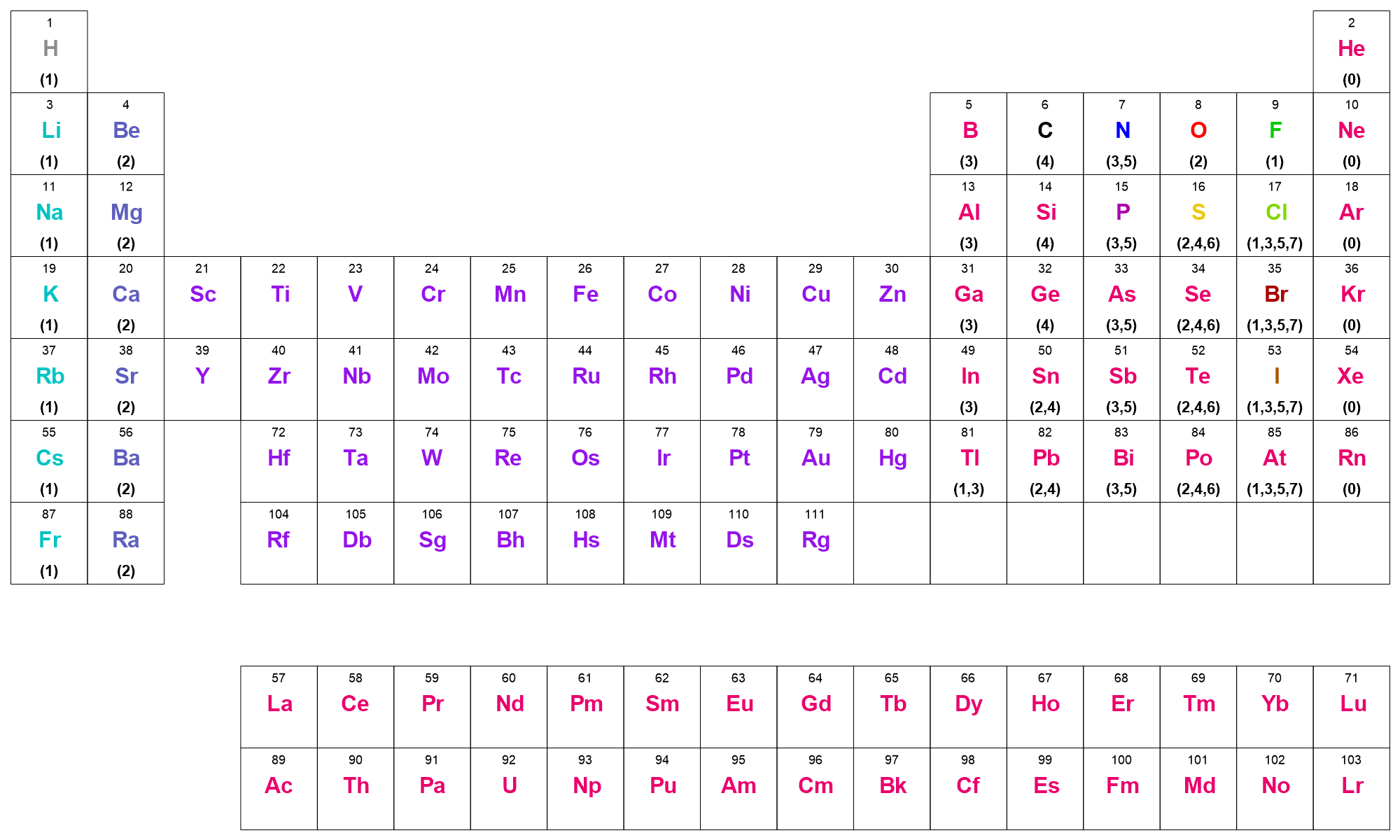
MDL Valence Model
See also
The following functions use the MDL valence model:
OEDefaultMDLHCountfunctionOEAssignMDLHydrogensfunctionOEMDLGetValencefunctionOEIsValidMDLAtomValence predicate
OpenEye Valence Model
OpenEye Charge Model
The OpenEye formal charge model assigns formal charges to elements based upon
their total valence.
In OEChem TK, this functionality is invoked by the
OEAssignFormalCharges function.
If the formal charge on an atom is non-zero, it is left unchanged.
- Hydrogen
If the valence isn’t one, the formal charge is +1.
- Boron
If the valence is four, the formal charge is +1.
- Carbon
If the valence is three, the formal charge is +1 if the atom has a polar neighbor, i.e. N, O or S, and formal charge -1 otherwise.
- Nitrogen
If the valence is two, the formal charge is -1, and if the valence is four the formal charge is +1.
- Oxygen
If the valence is one, the formal charge is -1, and if the valence is three the formal charge is +1.
- Phosphorous
If the valence is four, the formal charge is +1.
- Sulfur
If the valence is 1, the formal charge is -1, if the valence is three the formal charge is +1, if the valence is 5, the formal charge is -1, if the valence is four and the degree is four the charge is +2.
- Chlorine
If the valence is 0 the formal charge is -1, if the valence is four the formal charge is +3.
- Fluorine, Bromine, Iodine
If the valence is zero, the formal charge is -1.
- Magnesium, Calcium, Zinc
If the valence is zero, the formal charge is +2.
- Lithium, Sodium, Potassium
If the valence is zero, the formal charge is +1.
- Iron
If the valence is zero, the formal charge is +3 if the partial charge is 3.0, and +2 otherwise.
- Copper
If the valence is zero, the formal charge is +2 if the partial charge is 2.0, and +1 otherwise.
For the remaining elements, if the valence of an atom is zero, its formal charge is set from its partial charge.
OpenEye Hydrogen Count Model
OpenEye’s hydrogen count valence model is used by OEChem TK when neither hydrogen
counts nor valence are specified.
The typical uses are reading molecules from PDB or XYZ format files without explicit
hydrogens.
This functionality is invoked by
OEAssignImplicitHydrogens, which must
always be followed by a call to
OEAssignFormalCharges.
This valence model is unique in that it only partially updates hydrogen counts,
assuming the unfilled valences will be corrected by OpenEye’s charge valence
model above.
In MDL’s model for example, a neutral sodium atom is assumed to have one implicit hydrogen,
i.e. sodium hydride instead of sodium metal.
In OpenEye’s hydrogen count valence model, a disconnected sodium atom is assumed
to be a sodium cation, [Na+].
When reading from PDB files, this is a very reasonable assumption.
Note that although the OpenEye hydrogen count valence model often sets charge
and protonation states to physiological conditions, it is neither intended to
be a pKa nor ionization state predictor. Instead, it is a normalization.
Much like many registry systems and the MDL valence model which will convert
C(=O)[O-] to C(=O)O for registration purposes, this valence
model converts the opposite direction to C(=O)[O-].
Carbon is always assumed to be at least four valent, and therefore neutral.
Nitrogens that are conjugated (have double bonds, or have neighbors that have double bonds, in their Kekulé representations) are assumed at least three valent and neutral, whilst all other nitrogens are assumed to be (minimum) four valent, with a +1 formal charge.
Oxygens are assumed to be at least two valent and neutral, unless they have a single bond to an atom that is doubly bonded to oxygen, in which case it’s assumed to be one valent, with a -1 formal charge.
Sulfur is assumed to always be at least two valent.
All other elements are assumed to have no implicit hydrogens, and the formal charge as specified by the OpenEye charge model. This models all disconnected halogens as halide anions, and when disconnected the metals listed above as cations.
These rules are sufficient to reasonably protonate proteins read from PDB files. However, as described above, these rules are not intended to be a comprehensive rule-based pKa predictor. Users interested in predicting physiological ionization, and protonation/disassociation state enumeration should contact OpenEye, Cadence Molecular Sciences (http://www.eyesopen.com/) about our tools for exactly this task.