How to Guides for Spruce Floes¶
How can I provide Spruce information about the bound ligand?¶
In most cases, Spruce automatically identifies the bound ligand without user intervention. If the automated ligand detection fails, there are several parameters that the user can supply in order to assist Spruce:
Provide a set of ligand names (e.g. LIG, 5KV, etc.). There is also a strict option, in the event the user wants to only find the provided list of ligands, othwerwise the provided names are used in addition to the automated processes. Note, the ligand names are not allowed to be UNK or UNL, as these are reserved names in the PDB spec, where they have special meaning. In this event we recommend changing them in the PDB file.
Increase the number of atoms and number of residues, this is neccesary for larger peptidic ligands.
Provide a reference structure with a ligand in the binding site of interest.
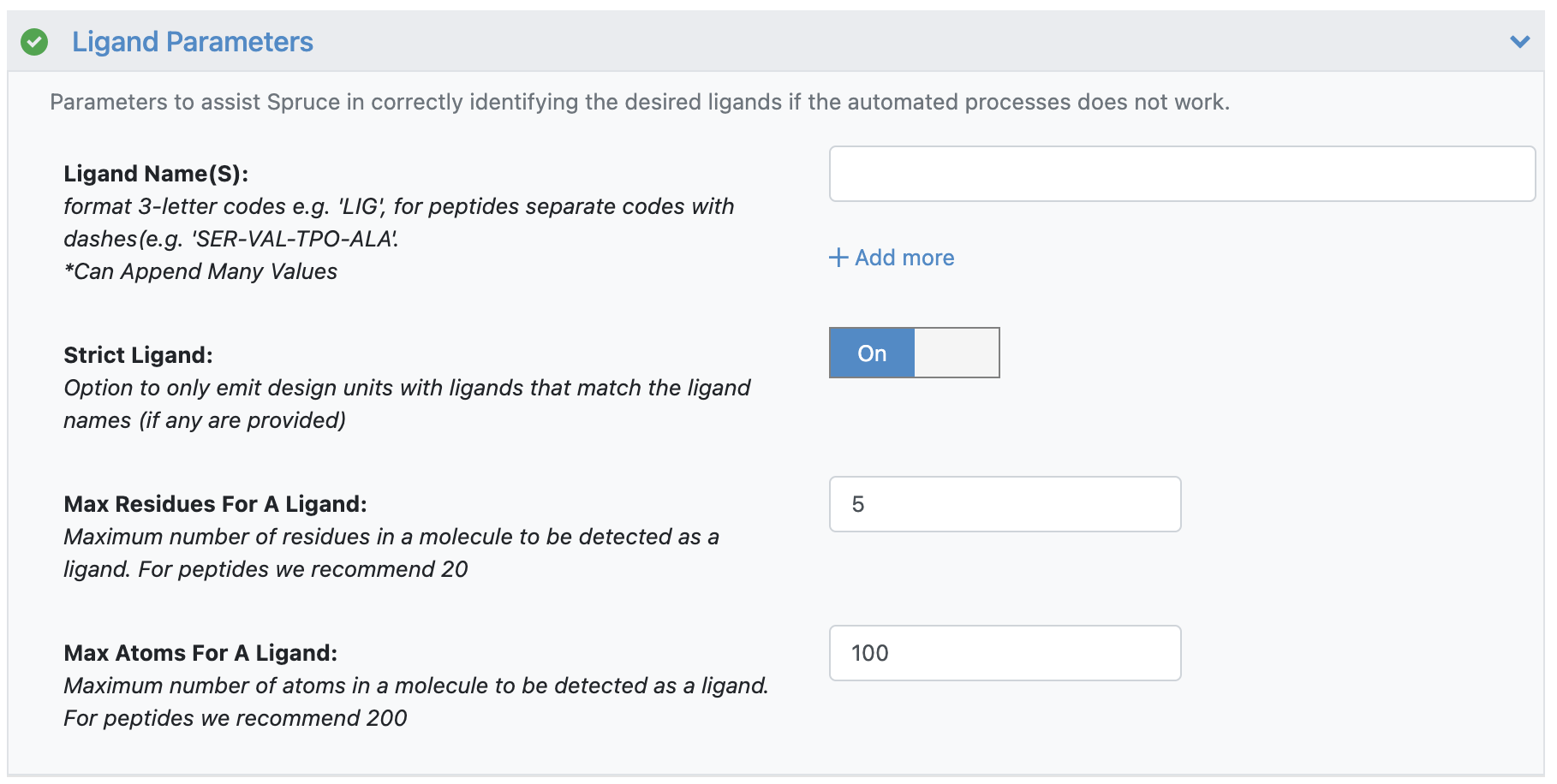
Ligand parameters(s)¶
How can I generate a prepared design unit for an un-liganded structure?¶
For apo structures or protein structures without a known binding site, the following steps can be taken:
Provide a reference design unit (DU) that identifies the binding site of interest.
Provide a single residue in the binding site of interest (e.g. “HIS:102: :A”).
Enumerate potential binding sites: in this case, the floe sends prepared biological units to a pocket finding cube that will enumerate potential binding pockets into individual prepared design units.
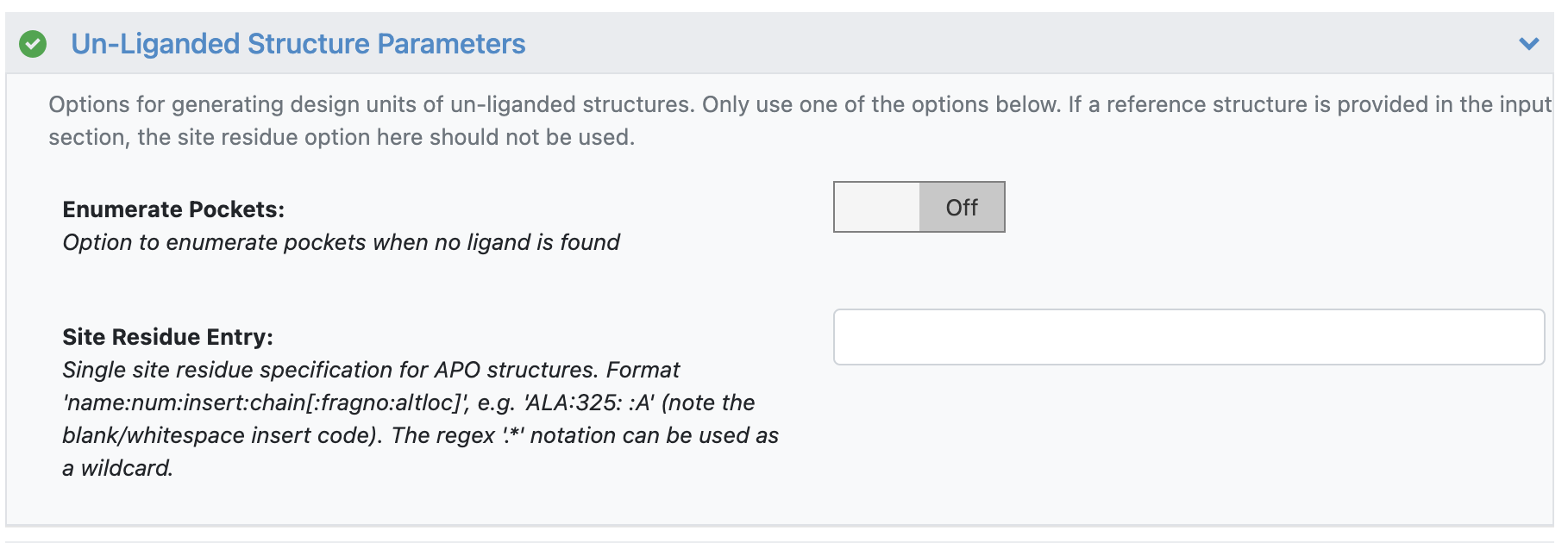
Unliganded parameters(s)¶
How do I edit a prepared design unit?¶
Once a design unit has been prepared by a Spruce floe, the dataset can be activated and shown in the 3D viewer. There are several options to edit different parts of the design unit. Guides to these steps can be found here: Protein editing.