Drawing a Molecule Surface
The 2D representation of the molecule surface is calculated by first drawing circles around each atom of the molecule and then identifying the external arc segments that define a continuous surface around the molecule. This process is illustrated in the table Table: Calculation of the 2D representation of a molecule surface
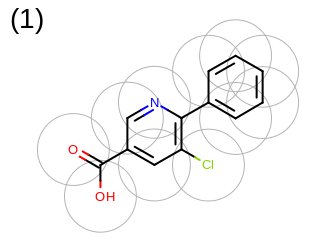
|
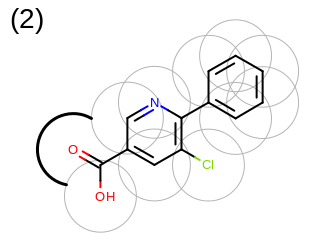
|
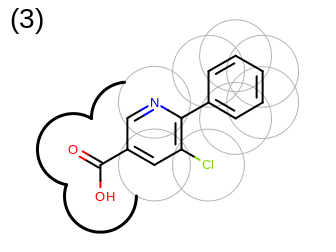
|
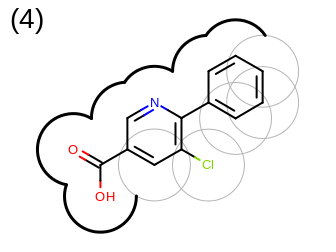
|
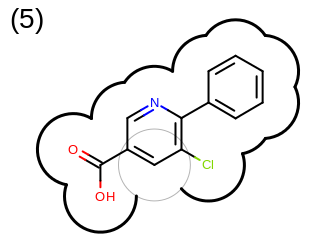
|
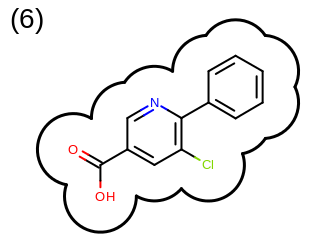
|
The following Listing 1 example shows how to
the display a molecule surface.
After creating an image object and preparing the molecule for 2D
depiction, an arc drawing functor is added to each atom of the
molecule by using the OESetSurfaceArcFxn
function.
The surface then can be drawn by calling the
OEDraw2DSurface function that calculates the
arc segments that form a continuous curve around the molecule.
The image created by Listing 1 is shown in
Figure: Example of surface drawing.
Listing 1: Example of surface drawing
OE2DMolDisplayOptions opts(width, height, OEScale::AutoScale);
opts.SetTitleLocation(OETitleLocation::Hidden);
opts.SetScale(OEGetMoleculeSurfaceScale(mol, opts));
OEEyelashArcFxn arcfxn(OEPen(OEGrey, OEGrey));
for (OEIter<OEAtomBase> atom = mol.GetAtoms(); atom; ++atom)
OESetSurfaceArcFxn(mol, atom, arcfxn);
OE2DMolDisplay disp(mol, opts);
OEDraw2DSurface(disp);
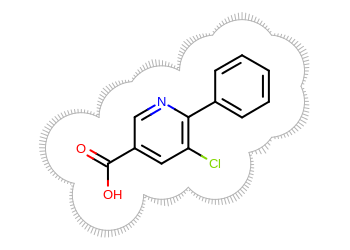
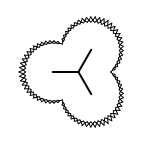
Example of surface drawing
The Listing 1 example uses the
OESurfaceArcStyle::Eyelash style to draw the
molecule surface.
The following table lists all surface drawing styles that are
available in Grapheme TK.
|
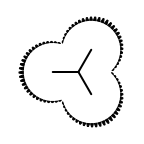
|
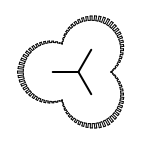
|
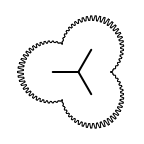
|
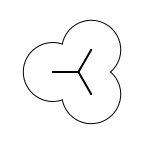
|
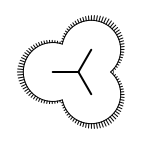
|
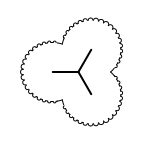
|
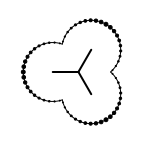
|
|
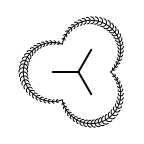
|
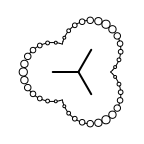
|
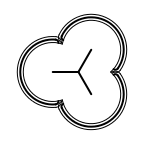
|
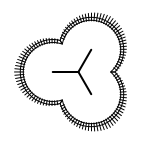
|
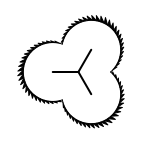
|
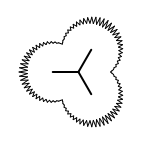
|
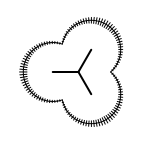
|
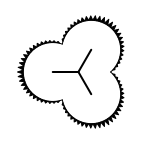
|
|
||
User-defined surface drawing functors can be implemented by deriving
from the OESurfaceArcFxnBase abstract base class.
In the Listing 2 example,
a user defined arc drawing functor is implemented that colors the
eyelash arcs according to the atom they belong to.
The image created by Listing 2 is shown in
Figure: Example of user-defined surface drawing.
Listing 2: Example of user-defined surface drawing
class AtomColorArcFxn : public OESurfaceArcFxnBase
{
public:
AtomColorArcFxn() {}
bool operator()(OEImageBase &image, const OESurfaceArc &arc) const
{
const OE2DAtomDisplay* adisp = arc.GetAtomDisplay();
if (adisp == nullptr || !adisp->IsVisible())
return false;
const OEColor color = adisp->GetLabelFont().GetColor();
const OEPen pen(color, color, OEFill::Off, 1.0);
const OE2DPoint center = arc.GetCenter();
const double radius = arc.GetRadius();
const double bgnAngle = arc.GetBgnAngle();
const double endAngle = arc.GetEndAngle();
OEDrawEyelashSurfaceArc(image, center, bgnAngle, endAngle, radius, pen);
return true;
}
OESurfaceArcFxnBase *CreateCopy() const
{
return new AtomColorArcFxn();
}
};
void Draw2DSurface(OEImageBase& image, OEMolBase& mol)
{
OEPrepareDepiction(mol);
OE2DMolDisplayOptions opts(image.GetWidth(), image.GetHeight(), OEScale::AutoScale);
opts.SetTitleLocation(OETitleLocation::Hidden);
opts.SetScale(OEGetMoleculeSurfaceScale(mol, opts));
AtomColorArcFxn arcfxn;
for (OEIter<OEAtomBase> atom = mol.GetAtoms(); atom; ++atom)
OESetSurfaceArcFxn(mol, atom, arcfxn);
OE2DMolDisplay disp(mol, opts);
OEDraw2DSurface(disp);
OERenderMolecule(image, disp);
}
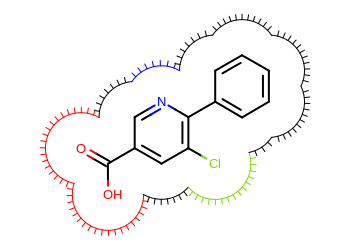
Example of user-defined surface drawing
The Listing 3 example shows
how to project atom properties, such as partial charges into the molecule
surface.
The image created by Listing 3 is shown in
Figure: Example of depicting atom properties.
Listing 3: Example of depicting atom properties on the molecule surface
class AtomPartialChargeArcFxn : public OESurfaceArcFxnBase
{
public:
AtomPartialChargeArcFxn(const OELinearColorGradient cg)
: m_colorg(cg)
{ }
bool operator()(OEImageBase &image, const OESurfaceArc &arc) const
{
const OE2DAtomDisplay* adisp = arc.GetAtomDisplay();
if (adisp == nullptr || !adisp->IsVisible())
return false;
const OEAtomBase* atom = adisp->GetAtom();
if (atom == nullptr)
return false;
const double charge = atom->GetPartialCharge();
if (charge == 0.0)
return true;
const OEColor color = m_colorg.GetColorAt(charge);
OEPen pen;
pen.SetForeColor(color);
pen.SetLineWidth(2.0);
const OE2DPoint center = arc.GetCenter();
const double radius = arc.GetRadius();
const double bAngle = arc.GetBgnAngle();
const double eAngle = arc.GetEndAngle();
const double edgeAngle = 5.0;
const unsigned dir = OEPatternDirection::Outside;
const double patternAngle = 10.0;
OEDrawBrickRoadSurfaceArc(image, center, bAngle, eAngle, radius, pen,
edgeAngle, dir, patternAngle);
return true;
}
OESurfaceArcFxnBase *CreateCopy() const
{
return new AtomPartialChargeArcFxn(m_colorg);
}
private :
OELinearColorGradient m_colorg;
};
void Draw2DSurfacePartialCharge(OEImageBase& image, OEMolBase& mol)
{
OEPrepareDepiction(mol);
OEMMFFAtomTypes(mol);
OEMMFF94PartialCharges(mol);
OE2DMolDisplayOptions opts(image.GetWidth(), image.GetHeight(), OEScale::AutoScale);
opts.SetTitleLocation(OETitleLocation::Hidden);
opts.SetScale(OEGetMoleculeSurfaceScale(mol, opts));
OEColorStop coloranion(-1.0, OEColor(OEDarkRed));
OEColorStop colorcation(+1.0, OEColor(OEDarkBlue));
OELinearColorGradient colorg(coloranion, colorcation);
colorg.AddStop(OEColorStop(0.0, OEColor(OEWhite)));
AtomPartialChargeArcFxn arcfxn(colorg);
for (OEIter<OEAtomBase> atom = mol.GetAtoms(); atom; ++atom)
OESetSurfaceArcFxn(mol, atom, arcfxn);
OE2DMolDisplay disp(mol, opts);
OEDraw2DSurface(disp);
OERenderMolecule(image, disp);
}
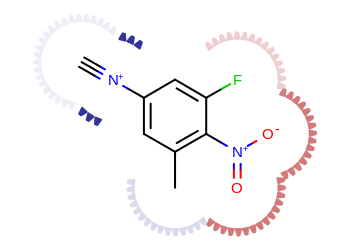
Example of depicting atom properties
See also
OELinearColorGradient class in the OEDepict TK manual
It is also possible to draw multiple 2D surfaces arc-by-arc.
In the Listing 4 example below the
arcs returned by the OEGet2DSurfaceArcs function
for the given atom display and radius are directly rendered below
the molecule diagram.
In this case it is important to call the OEGetMoleculeSurfaceScale
function with the largest radius scale of the 2D surfaces drawn.
This reduces the scaling of the molecule in order to able to fit the
molecule diagram and all of the arcs of the 2D surfaces into the image.
The image created by Listing 4 is shown in
Figure: Example of drawing multiple 2D surfaces.
Listing 4: Example of drawing multiple 2D surfaces
void Draw2DSurface(OE2DMolDisplay& disp, const OEUnaryPredicate<OEAtomBase>& atompred,
double radius, const OEColor& color)
{
OEPen penA(color, color, OEFill::Off, 2.0);
OEDefaultArcFxn arcfxnA(penA);
OEPen penB(OELightGrey, OELightGrey, OEFill::Off, 2.0, OEStipple::ShortDash);
OEDefaultArcFxn arcfxnB(penB);
OEImage& layer = disp.GetLayer(OELayerPosition::Below);
for (OEIter<OE2DAtomDisplay> ai = disp.GetAtomDisplays(); ai; ++ai)
{
const OE2DAtomDisplay& adisp = (*ai);
for (OEIter<const OESurfaceArc> arc = OEGet2DSurfaceArcs(disp, adisp, radius); arc; ++arc)
{
if (atompred(*adisp.GetAtom()))
arcfxnA(layer, arc);
else
arcfxnB(layer, arc);
}
}
}
void Draw2DSurfaces(OEImageBase& image, OEMolBase& mol)
{
OEPrepareDepiction(mol);
OE2DMolDisplayOptions opts(image.GetWidth(), image.GetHeight(), OEScale::AutoScale);
opts.SetTitleLocation(OETitleLocation::Hidden);
opts.SetScale(OEGetMoleculeSurfaceScale(mol, opts, 1.50));
OE2DMolDisplay disp(mol, opts);
Draw2DSurface(disp, OEHasAtomicNum(OEElemNo::C), 1.00, OEBlack);
Draw2DSurface(disp, OEHasAtomicNum(OEElemNo::N), 1.25, OEDarkBlue);
Draw2DSurface(disp, OEHasAtomicNum(OEElemNo::O), 1.50, OEDarkRed);
OERenderMolecule(image, disp);
}
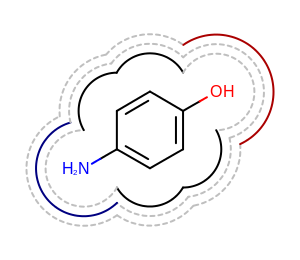
Example of drawing multiple 2D surfaces
See also
OESurfaceArcScale namespace
The last Listing 5 example shows how to
draw a 2D surface with various radii.
It draws two surfaces one with a minimum radius scale and one with various radius
depending on the covalent radius of the atoms.
The OEGetMoleculeSurfaceScale function has to be called
in this example too with the largest radius scale in order to able to fit the molecule diagram
and all of the arcs of the 2D surfaces into the image.
The image created by Listing 5 is shown in
Figure: Example of drawing 2D surface with various radii.
Listing 5: Example of drawing 2D surface with various radii
void DrawSurfaces(OEImageBase& image, OEMolBase& mol)
{
OEAssignCovalentRadii(mol);
const double minradius = OEGetCovalentRadius(OEElemNo::H);
double maxrscale = -DBL_MAX;
std::vector<double> radiusScales(mol.GetMaxAtomIdx(), 0.0);
for (OEIter<const OEAtomBase> atom = mol.GetAtoms(); atom; ++atom)
{
const double rscale = (atom->GetRadius()-minradius) + OESurfaceArcScale::Minimum;
radiusScales[atom->GetIdx()] = rscale;
maxrscale = std::max(maxrscale, rscale);
}
OE2DMolDisplayOptions opts(image.GetWidth(), image.GetHeight(), OEScale::AutoScale);
opts.SetTitleLocation(OETitleLocation::Hidden);
opts.SetScale(OEGetMoleculeSurfaceScale(mol, opts, maxrscale));
OE2DMolDisplay disp(mol, opts);
OEImage& layer = disp.GetLayer(OELayerPosition::Below);
const OEPen penA(OELightGrey, OELightGrey, OEFill::Off, 2.0, OEStipple::ShortDash);
const OEDefaultArcFxn arcfxnA(penA);
for (OEIter<OESurfaceArc> arc = OEGet2DSurfaceArcs(disp, OESurfaceArcScale::Minimum); arc; ++arc)
arcfxnA(layer, arc);
const OEPen penB = OEPen(OEGrey, OEGrey, OEFill::Off, 2.0);
const OEDefaultArcFxn arcfxnB(penB);
for (OEIter<OESurfaceArc> arc = OEGet2DSurfaceArcs(disp, radiusScales); arc; ++arc)
arcfxnB(layer, arc);
OERenderMolecule(image, disp);
}
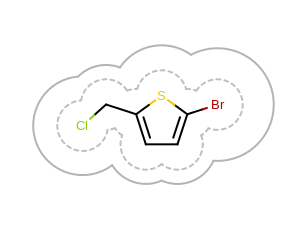
Example of drawing 2D surface with various radii
See also
OEGetCovalentRadius function in the OEChem TK manual