OEPrepareMultiAlignedDepiction
bool OEPrepareMultiAlignedDepiction(OEChem::OEMolBase &fitmol,
const OEChem::OEMolBase &refmol,
OESystem::OEIter<const OEChem::OEMatchBase> &miter)
bool OEPrepareMultiAlignedDepiction(OEChem::OEMolBase &fitmol,
const OEChem::OEMolBase &refmol,
OESystem::OEIterBase<const OEChem::OEMatchBase> *miter)
Generates the 2D coordinates of a molecule by maximizing its overlay with a reference 2D molecule based on multiple overlaps between the two molecules.
- fitmol
The molecule being fitted to be aligned to the reference molecule.
- refmol
The reference molecule of which 2D coordinates are remain unchanged.
- miter
The iterator that stores the matches on which the molecule alignment is based on.
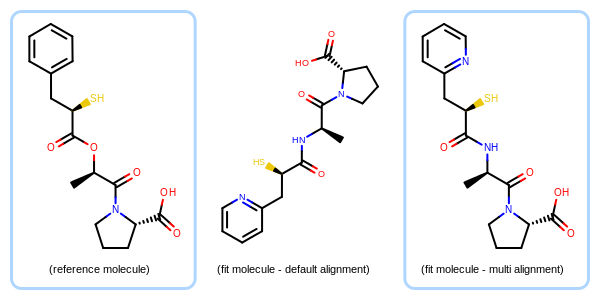
Example of the molecule alignment based on multiple fragments
(left) - depiction of a reference molecule (middle) - default depiction of a fit molecule (right) - depiction of the fit molecule aligned to the reference based on multiple fragments
Hint
The overlap between two molecules can be generated by the
OEGetFPOverlap function of GraphSim TK.
The OEGetFPOverlap returns matched fragments
of two molecules that are considered equivalent based on a specific
fingerprint type.
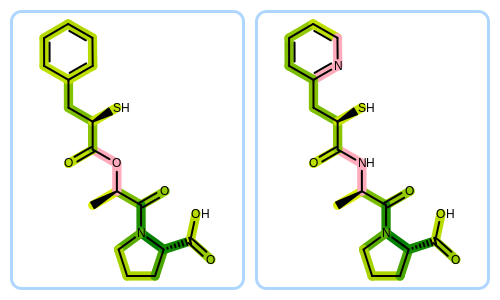
The Python script that visualizes molecule similarity based on fingerprints can be downloaded from the OpenEye Python Cookbook
See also
Molecule Alignment chapter
Fingerprint Overlap chapter in GraphSim TK
OEPrepareAlignedDepictionfunctionListing 3example code in Molecule Alignment chapter