Release Highlights 2021.1
OEDocking TK: Receptors in Design Unit
OEReceptor as an integral part of an OEDesignUnit,
with properly prepped structures and contained in an OEDU file, is fully integrated
in all workflows. Receptors in an OEDesignUnit can now be created using the
Make Receptor GUI application, as well as the SPRUCE and ReceptorInDU command-line applications.
The Make Receptor GUI application has been extended
to be a fully functional DesignUnit builder in addition to building receptors. A new
DesignUnit can be created from a PDB/MMCIF file (with or without an associated
MTZ file), or by combining molecules from various molecule files of any format.
SPRUCE options are exposed through a graphical interface giving
users control over the level of preparation the structure should go through.
Existing receptors, both in OEDU or OEB format, can also be opened and edited
as desired in Make Receptor.
OEMakeReceptor now automatically detects protein constraints.
The detected protein constraints are kept disabled by default, and can be completely
turned off if desired,
functionality that was previously only available in the Make Receptor application.
Both the detected constraints and any custom constraints
can be enabled and edited via the OEMakeReceptor
interface, or using commands from OEReceptorProteinConstraint and
OEReceptorCustomConstraints classes in the toolkit.
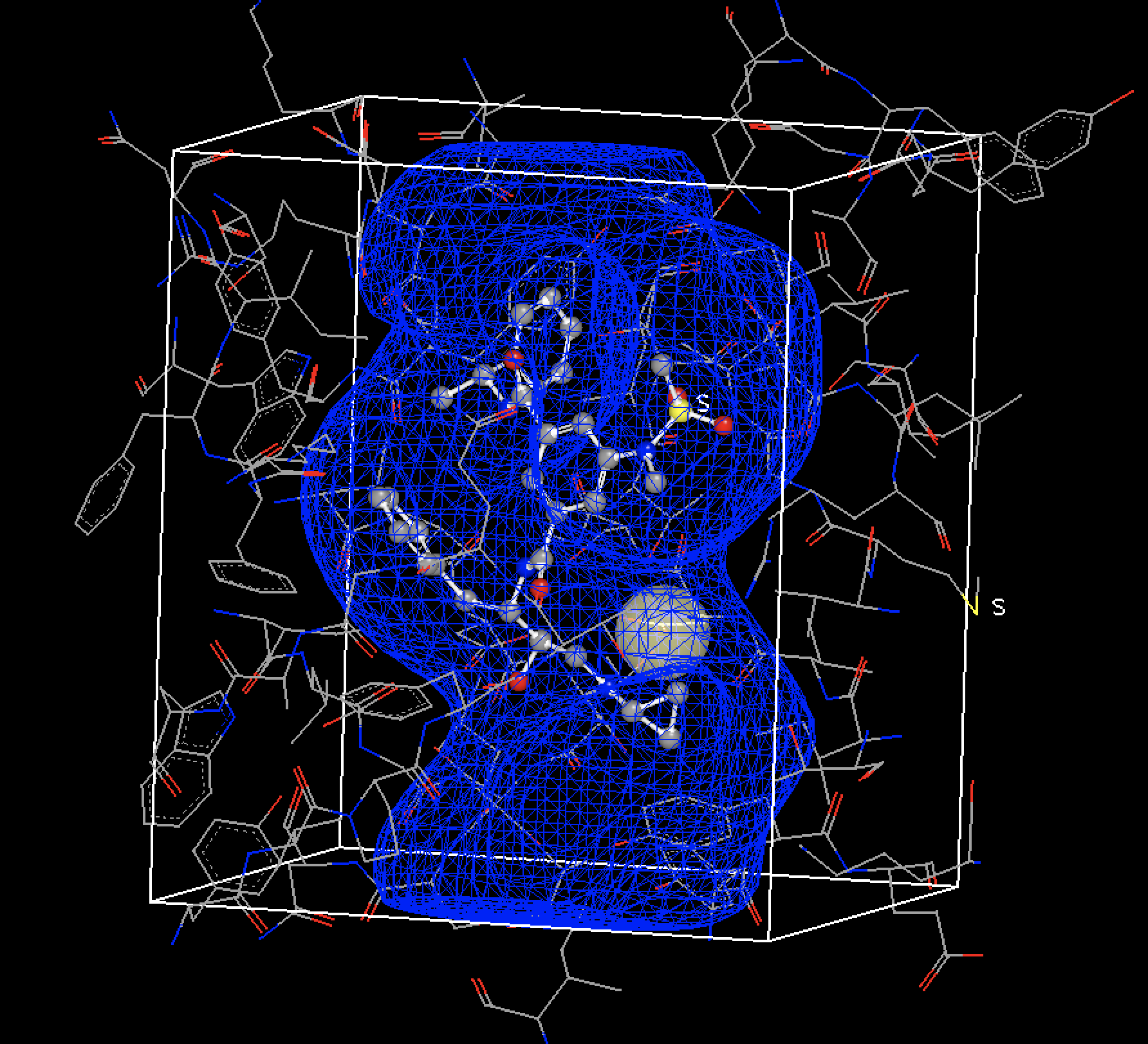
OEDocking receptor for the 3TPP BACE1 Complexed with Inhibitor with target structure, bound ligand, and a protein constraint. Displayed in the Make Receptor viewer.
Having the receptors as part of the OEDesignUnit with properly prepped structures makes
it easier for the docked and posed structures to be used in further downstream modeling,
which is especially necessary when working with protein force fields like FF14SB.
The new receptors with properly prepped protein also enabled use of modern
force fields like FF14SB and Parsley for structure optimization
in flexible OEPosit.
OESiteHopper: Application Suite for Rapid Protein Binding Site Comparison
The 2021.1 release introduces SiteHopper, which can search a database
containing hundreds of thousands of proteins in a few minutes for
proteins with similar binding sites to a query protein. SiteHopper will
output the most similar biomolecules, overlaid by binding site, for visualization
and analysis. A visual representation of the binding site is also output.
SiteHopper can also create databases for searching from a series of biomolecular
structures in OEDU format.
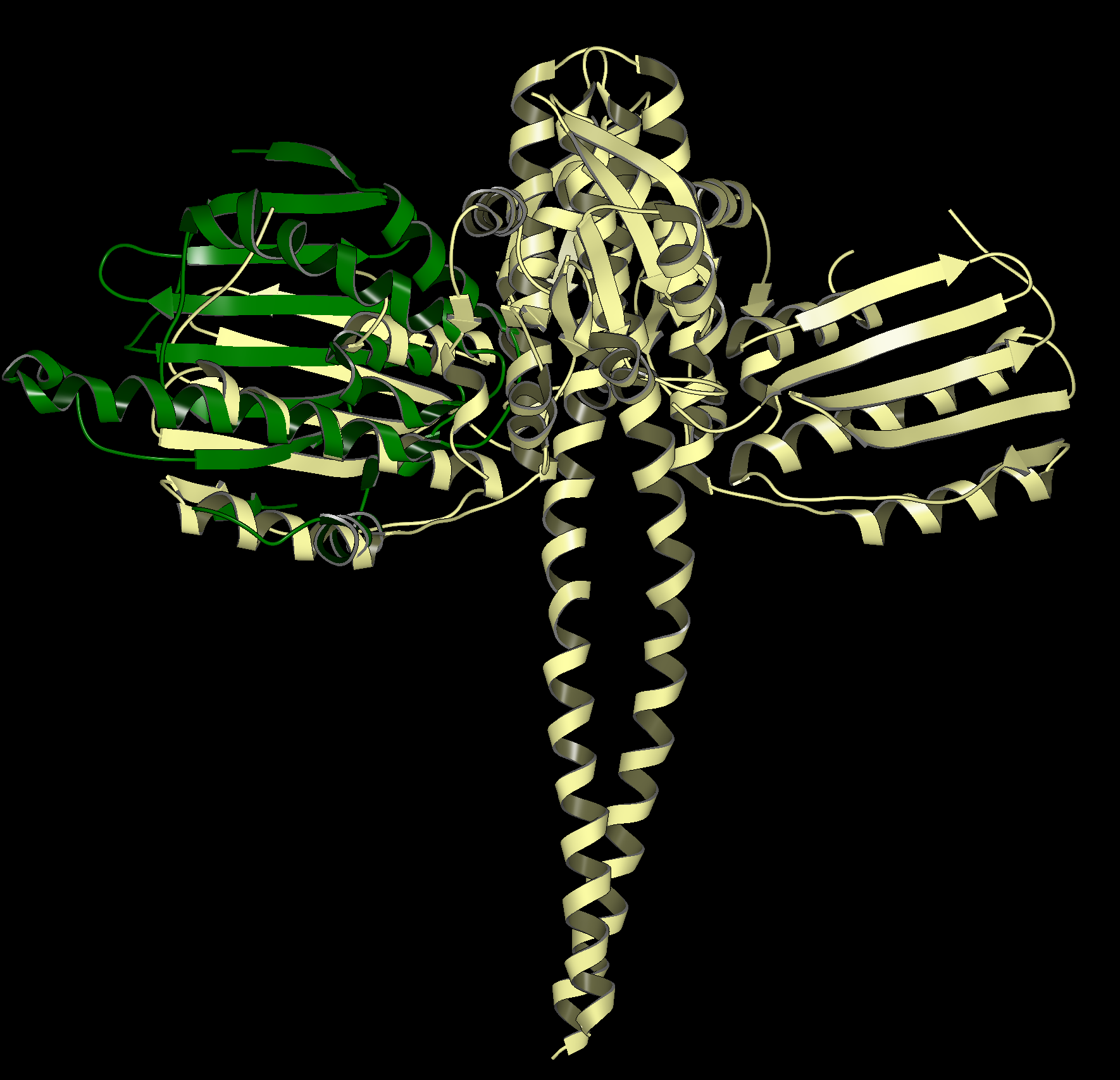
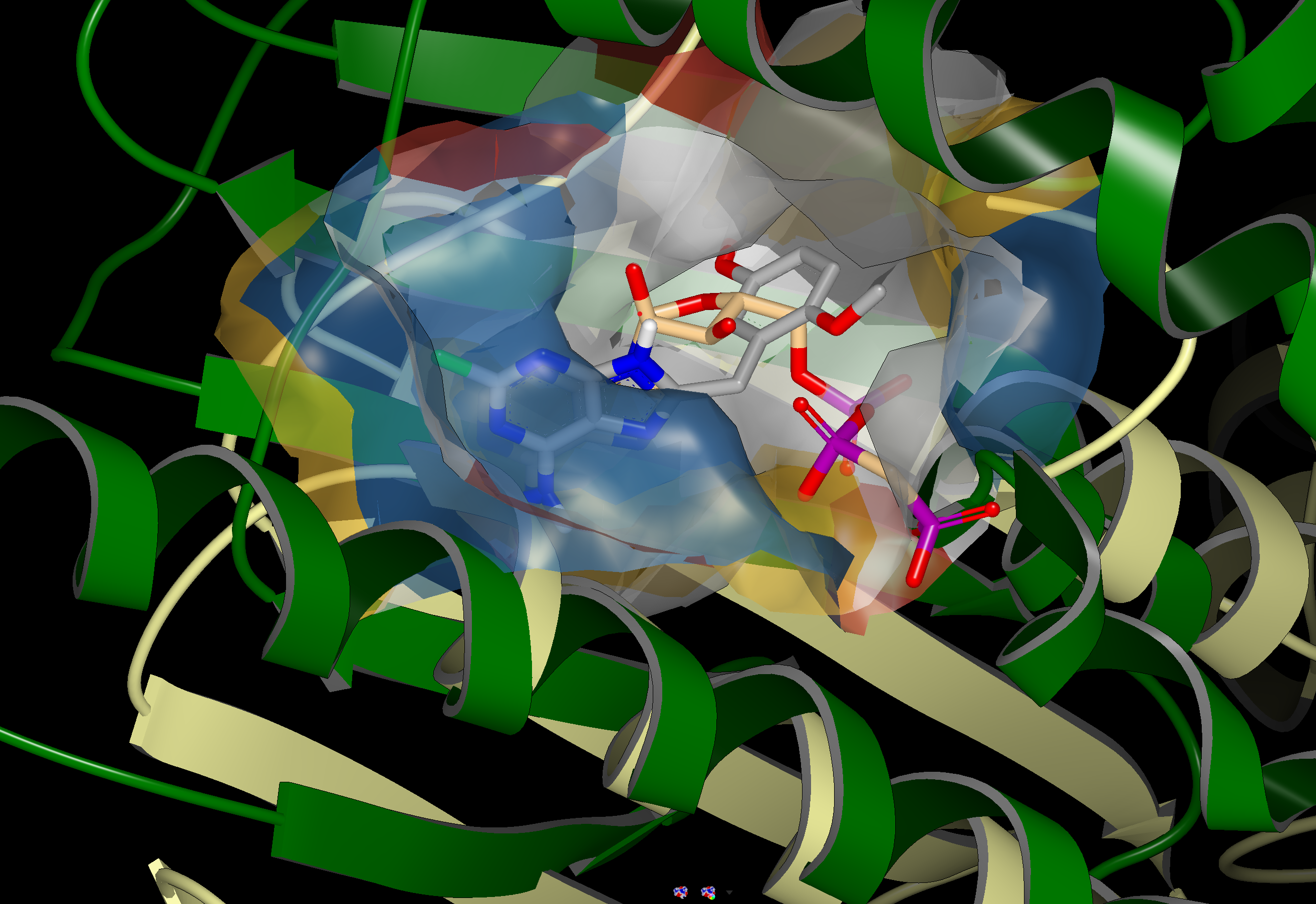
SiteHopper finds proteins with similar binding sites which searching by sequence similarity would overlook. An example is shown in the figure below. 1UYG is a structure of human heat shock protein 90-alpha, bound to 8-(2,5-dimethoxy-benzyl)-2-fluoro-9H-purin-6-ylamine. 5IUN is a structure of the DesK-DesR complex, bound to AMP-PCP.
The image on the left shows the overlay of 1UYG (green) and 5UIN (light yellow), zoomed out to show the major structural differences between the two proteins. The image on the right zooms in on the binding sites, with a surface showing the type of residues present in each binding site. Blue represents acidic, red represents basic, yellow represents polar, and white represents non polar. Despite a sequence similarity of only 46%, 1UYG and 5IUN may be targetable by similar ligands, as they have very similar steric and electrostatic properties in their binding sites.
|
|
1UYG human hsp 90 (green) overlaid with 5UIN N-formyltransferase (yellow), full struture view (left) and binding site view (right).
VIDA 5: Major Update and OEDesignUnit Support
VIDA has undergone major changes including:
Upgrade to Python3.
Upgrade to Qt5 with PySide2 providing for user-interface improvements.
Updated to run on the same platforms as OpenEye Toolkits and Applications.
This allows us to release VIDA along side the toolkits and applications in our twice-yearly releases.
In addition, VIDA now has support for MMCIF files and improvements to I/O from other file format types. VIDA can also read prepared protein structures in OEDesignUnit format produced with SPRUCE and Make Receptor along with the receptor objects that are now stored on these design units.
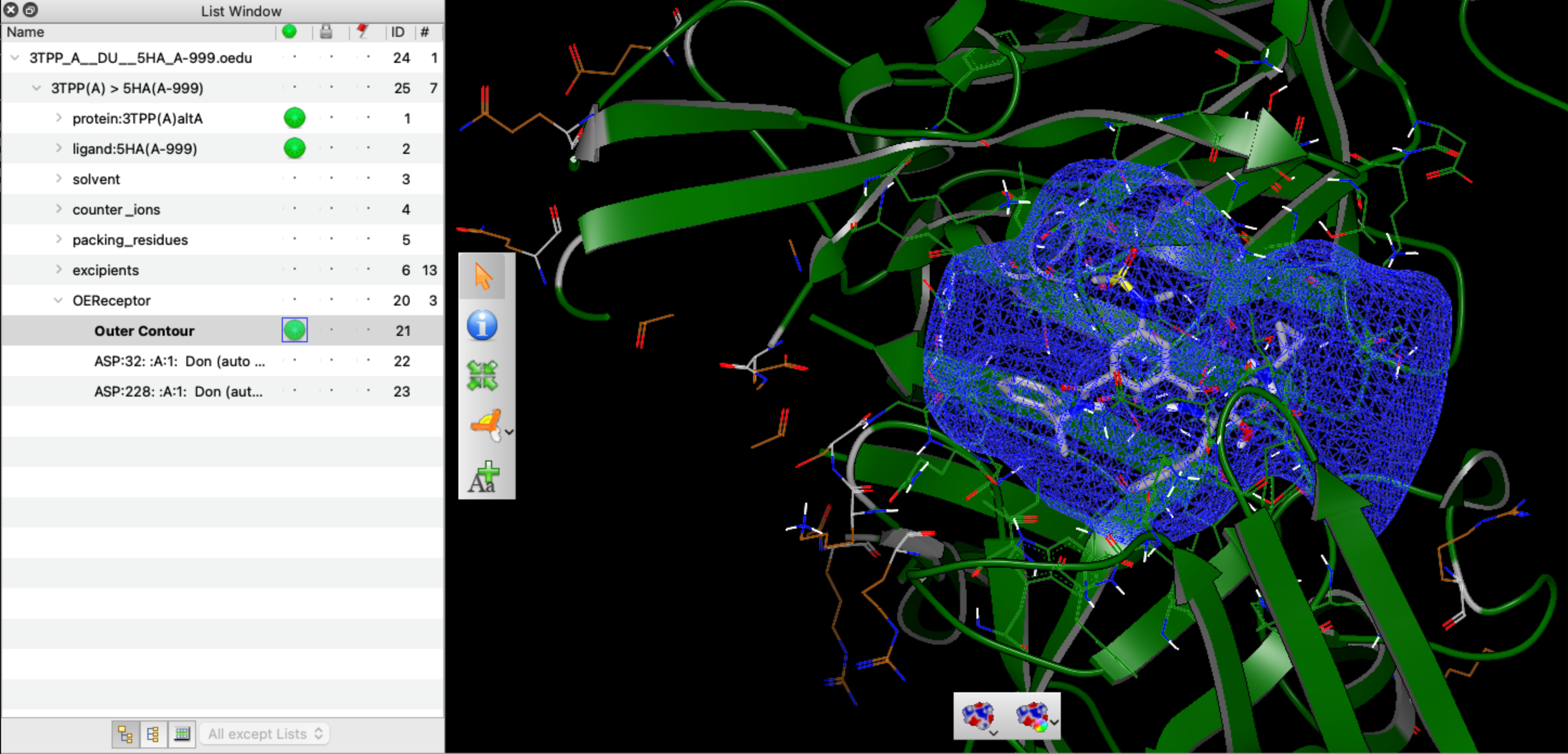
VIDA DesignUnit Support with receptors showing the prepared beta-secretase structure 3TPP.
Supported Platforms
Package
Versions
Linux
Windows
macOS
Python
3.7, 3.8, 3.9
RHEL7/8, Ubuntu18/20
Win10
10.14, 10.15, 11
C++
RHEL7/8, Ubuntu18/20
Win10 (VS2017, VS2019)
10.14, 10.15, 11
Java
1.8, 11
RHEL7/8, Ubuntu18/20
Win10
10.14, 10.15, 11
C#
Win10 (VS2017, VS2019)
General Notices
C++ CUDA-enabled toolkits and features are now supported on RHEL8 and Ubuntu20.
Support for GCC 9.x has been added. Support for GCC 4.x and GCC 5.x has been dropped.
Support for Python 3.6 has been dropped.
Support for VS2015 has been dropped.
Support for MacOS 11 has been added. Support for MacOS 10.13 has been dropped.
OpenEye applications and toolkits have not been optimized for the M1 Mac but run under Rosetta 2.