OEToolkits 2015.Jun
PDB Splitting
OEBio TK includes a powerful new API for automatically classifying and separating the various components of a macromolecular complex by their functional roles. This new API makes it easy to excise a ligand or protein/cofactor complex automatically or identify binding-site residues. The process is also fully customizable to allow internal definitions of the ligand to augment the splitting algorithm. For more information, please see the Splitting Macromolecular Complexes documentation.
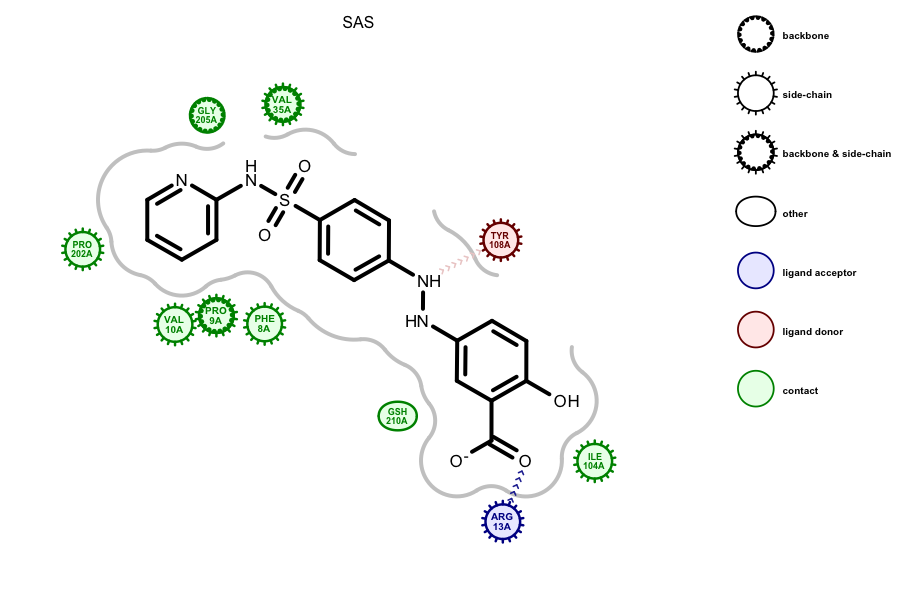
The above Sulfasalazine ligand was automatically identified in the
glutathione S-transferase binding site by using the
OESplitMolComplex function. The image was created
by running the
complex2img Grapheme
TK example on the 13GS PDB entry. Previous versions
required an external definition of the ligand; this new version
bypasses that requirement for ease of use.
PAINS Filter
Pan Assay Interference Compounds (PAINS) filter was added to OEMolProp TK by adapting the supplementary material from the original [Baell-2010] paper. The PAINS filter can be used as a prefilter set for screening or, more interestingly, as a post-processing filter to identify possible protein-reactive or other assay-confounding functionalities within hits. Any hits so identified will require more care to determine if the measured activity is valid. The PAINS filters can give valuable insights into lead optimization priorities.
Matched Molecular Pair Improvements
There is a wealth of information stored in a chemical series. Extracting this property change information from chemical analogs is at the heart of a matched molecular pair analysis. One challenge is capturing molecular differences corresponding to substitution of a hydrogen with a larger group in an unsupervised analysis approach. This new indexing capability generates significantly more matched molecular pairs during the analysis and yields additional information about substituent effects on molecular properties. More matched pairs means more information is being extracted from the chemical series.
This next beta release of OEMedChem TK improves the matched molecular pair analysis engine to robustly and efficiently support the identification of molecular pairs due to differences in hydrogen-atom substitutions:
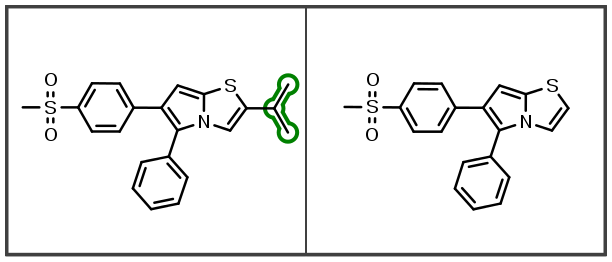
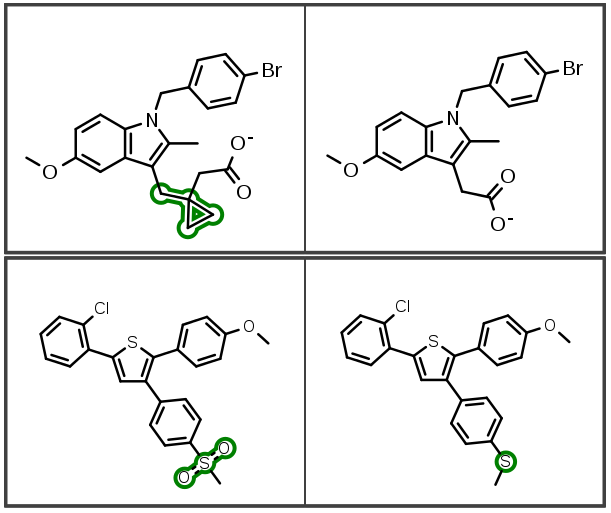
Additionally, indexing improvements also identify new linker insertions and modifications:
Custom Ring Template Dictionaries
Depiction of complex molecules in 2D is both a science and an art. For this reason, OEChem TK can now utilize custom ring template dictionaries specified by the user. This allows OpenEye Toolkits to adapt immediately to changing project demands. OpenEye will gladly continue to incorporate ring templates submitted to us, but this new feature will be more flexible. The Custom Ring Template Dictionary Guide explains the tools and process to start using custom internal dictionaries.
Anaconda Support
We want to work where you do. At OpenEye, we support all major platforms and Continuum’s Anaconda is becoming a major platform for scientific Python. Anaconda is a Python distribution that installs commonly used scientific packages by default. OpenEye Python Toolkits have added binary support for Anaconda.
Note
Using the PIP installation mechanism described below greatly simplifies this compatibility; the user should not even notice that Anaconda Python requires a different binary.
One-Command Python Installation
OpenEye Python Toolkits can now be installed with one command:
$ pip install -i https://pypi.binstar.org/OpenEye/simple OpenEye-toolkits
...
$ oecheminfo.py
Installed OEChem version: |oechemversion| platform: ubuntu-14.04-g++4.8-x64 built: |builddate|
...
See the Quick Start guide to learn how to make this command even shorter:
pip install OpenEye-toolkits
The command above will automatically select the correct version and operating system in a way that is natural for any pure Python project.
The Python toolkit package name has been changed from
OpenEyetoOpenEye-toolkits. This will change how the previous toolkit is uninstalled from avirtualenv.examples,docexamples, andtestsare now found under theopeneyemodule in theOpenEye-toolkitspackage. This allows examples and integration testing to be performed easily after installing theOpenEye-toolkitspackage like a proper Python package. Run the following command to execute the integration tests:(OpenEye)$ python -m openeye.examples.openeye_tests
In addition to OpenEye examples being placed in the Python
binorScriptsdirectory, examples can also be run through the Python interpreter’s-moption:(OpenEye)$ python -m openeye.examples.oechem.oecheminfo
py2exe should now work. OpenEye Python Toolkits will no longer use modules that are blacklisted by py2exe.
General Notices
This is the last release to support Visual Studio 2010 for C++.
This is the last release to support CMake 2.x in the C++ distributions; all future releases will use CMake 3.1+. The CMake files shipped with the C++ toolkits have been made compatible in both CMake 2.x and CMake 3.1+, but compatibility with CMake 2.x will not be tested after this release.