OEAddComplexSurfaceArcs
bool OEAddComplexSurfaceArcs(OEChem::OEMolBase &ligand,
const OEChem::OEMolBase &protein,
const OESurfaceArcFxnBase &solventExposedFxn=OEDefaultSolventArcFxn(),
const OESurfaceArcFxnBase &buriedFxn=OEDefaultBuriedArcFxn(),
const OEComplexSurfaceArcFxnBase &cavityFxn=OEDefaultCavityArcFxn(),
const OEComplexSurfaceArcFxnBase &voidFxn=OEDefaultVoidArcFxn())
Takes a ligand-protein complex and generates the 2D molecule surface of the ligand, where the arcs of the surface are annotated based on the distance calculated between the accessible surface of the ligand and the protein. See Figure: Example of surface drawing with the OEAddComplexSurfaceArcs function.
- ligand
The molecule for which the 2D surface is generated.
- protein
The macromolecule whose contact with the ligand influences the surface drawing.
- solventExposedFxn
This functor is used to draw those arc segments of the 2D molecule surface where the ligand is exposed to the solvent in the 3D complex. By default, the OEDefaultSolventArcFxn functor is used.
- buriedFxn
This functor is used to draw those arc segments of the 2D molecule surface where the ligand and the protein tightly fit in the 3D complex. By default, the OEDefaultBuriedArcFxn functor is used.
- cavityFxn
This functor is used to draw those arc segments of the 2D molecule surface where there is a cavity detected between the ligand and the protein in the 3D complex. By default, the OEDefaultCavityArcFxn functor is used.
- voidFxn
This functor is used to draw those arc segments of the 2D molecule surface where there is a small gap detected between the ligand and the protein in the 3D complex. By default, the OEDefaultVoidArcFxn functor is used.
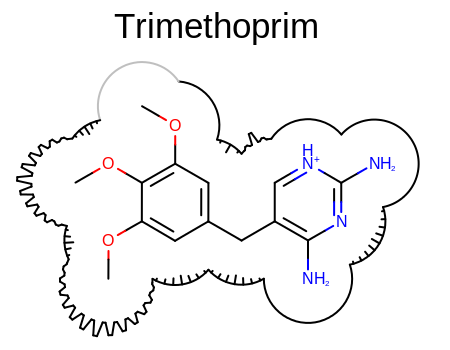
Example of surface drawing with the OEAddComplexSurfaceArcs function
See also