User-defined Fingerprint
The previous Fingerprint Generation chapter showed how to create circular, path and tree fingerprints with default parameters. These default parameters are calibrated on the Briem-Lessel [Briem-Lessel-2000], Hert-Willett [Hert-Willett-2004] and Grant [Grant-2006] benchmarks.
However, the GraphSim TK also provides facilities to construct user-defined fingerprints. When constructing a user-defined fingerprint, the following parameters have to be considered:
Atom and bond typing that define which atom and bond properties are encoded into the fingerprints (see the Atom and Bond Typing section)
Size of the fragments that are exhaustively enumerated during the fingerprint generation (see the Fragment Size section)
Size of the generated fingerprint (in bits) (see the Fingerprint Size section)
The following code snippet shows how to generate a 1024 bit long
fingerprint that encodes paths from 0 up to 5 bonds in length
with default atom and bond properties defined by the
OEFPAtomType.DefaultAtom and
OEFPBondType.DefaultBond constants,
respectively.
uint numbits = 1024;
uint minbonds = 0;
uint maxbonds = 5;
OEGraphSim.OEMakePathFP(fp,
mol,
numbits,
minbonds,
maxbonds,
OEFPAtomType.DefaultAtom,
OEFPBondType.DefaultBond);
Warning
Two fingerprints which are generated with different parameters will have different fingerprint types!
In Listing 14, two fingerprints are generated with
different parameters, namely they have a different number of bits.
This means that they also have different types, therefore,
no similarity value can be calculated between them.
Listing 14: Example of different path fingerprint types
OEFingerPrint fpA = new OEFingerPrint();
uint numbits = 1024;
uint minbonds = 0;
uint maxbonds = 5;
OEGraphSim.OEMakePathFP(fpA,
mol,
numbits,
minbonds,
maxbonds,
OEFPAtomType.DefaultAtom,
OEFPBondType.DefaultBond);
OEFingerPrint fpB = new OEFingerPrint();
numbits = 2048;
OEGraphSim.OEMakePathFP(fpB,
mol,
numbits,
minbonds,
maxbonds,
OEFPAtomType.DefaultAtom,
OEFPBondType.DefaultBond);
Console.WriteLine("same fingerprint types = " + OEGraphSim.OEIsSameFPType(fpA, fpB));
Console.WriteLine("{0:0.000}", OEGraphSim.OETanimoto(fpA, fpB));
The output of Listing 14 is the following:
same fingerprint types = False
Fatal: fingerprint type mismatch!
Atom and Bond Typing
Listing 15 shows how to generate fingerprints for
two molecules with various atom and bond types
(depicted in Example molecules).
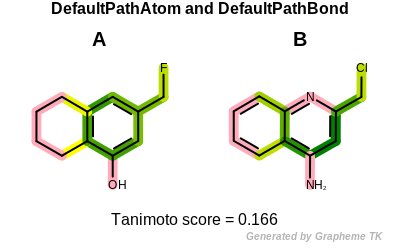
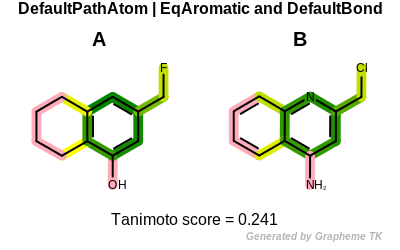
Reducing the number of atom and bond properties increases the similarity
between the two molecules (i.e. their
Tanimoto similarity).
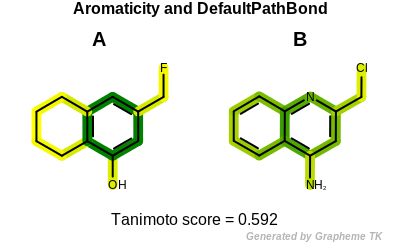
At the end, when only the topology of two molecules is considered,
i.e., whether or not their atoms and bonds belong to any ring system,
the fingerprints of the two molecules become identical.
These effects are illustrated in Table: Examples of depiction molecule similarity based on fingerprints.
See also
Visualizing Molecule Similarity section
Listing 15: Similarity calculation with various atom/bond typing
public class FPAtomTyping
{
public static int Main(string[] args)
{
OEGraphMol molA = new OEGraphMol();
OEChem.OESmilesToMol(molA, "Oc1c2c(cc(c1)CF)CCCC2");
OEGraphMol molB = new OEGraphMol();
OEChem.OESmilesToMol(molB, "c1ccc2c(c1)c(cc(n2)CCl)N");
PrintTanimoto(molA, molB, OEFPAtomType.DefaultAtom, OEFPBondType.DefaultBond);
PrintTanimoto(molA, molB, OEFPAtomType.DefaultAtom | OEFPAtomType.EqAromatic, OEFPBondType.DefaultBond);
PrintTanimoto(molA, molB, OEFPAtomType.Aromaticity, OEFPBondType.DefaultBond);
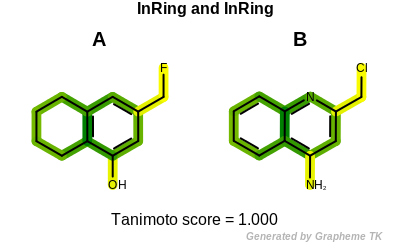
PrintTanimoto(molA, molB, OEFPAtomType.InRing, OEFPBondType.InRing);
return 0;
}
private static void PrintTanimoto(OEMolBase molA, OEMolBase molB, uint atype, uint btype)
{
OEFingerPrint fpA = new OEFingerPrint();
OEFingerPrint fpB = new OEFingerPrint();
uint numbits = 2048;
uint minb = 0;
uint maxb = 5;
OEGraphSim.OEMakePathFP(fpA, molA, numbits, minb, maxb, atype, btype);
OEGraphSim.OEMakePathFP(fpB, molB, numbits, minb, maxb, atype, btype);
Console.WriteLine("Tanimoto(A,B) = {0:0.000}", OEGraphSim.OETanimoto(fpA, fpB));
}
}
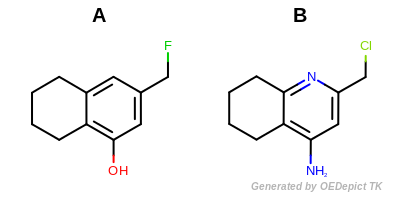
Example molecules
The output of Listing 15 is the following:
Tanimoto(A,B) = 0.166
Tanimoto(A,B) = 0.241
Tanimoto(A,B) = 0.592
Tanimoto(A,B) = 1.000
|
|
|
|
Table: Atom typing options and Table: Bond typing options list the currently available typing options.
atom typing constant |
encoded atom property |
|---|---|
bond typing constant |
encoded bond property |
|---|---|
See also
OEFPAtomTypenamespaceOEFPBondTypenamespace
Fragment Size
Circular, path and tree-based fingerprint generation involves molecular graph traversal to identify all unique radial, linear or branched fragments, respectively. When a path or tree fingerprint is initialized, the minimum and maximum number of bonds of the fragments that are encoded into the fingerprint can be specified. See Figure: Example of enumerated path fragments with increasing number of bonds and Figure: Example of enumerated tree fragments with increasing number of bonds.
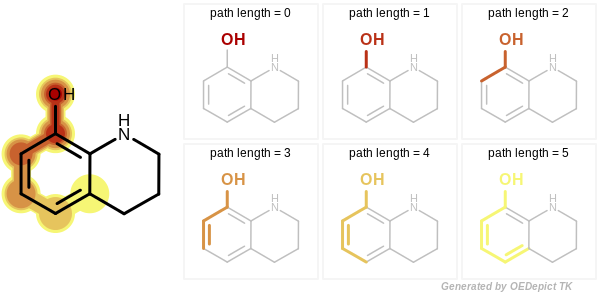
Example of enumerated path fragments with increasing number of bonds
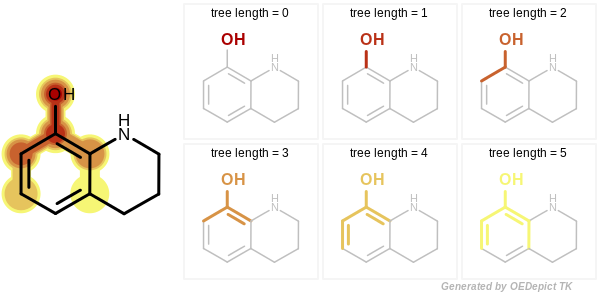
Example of enumerated tree fragments with increasing number of bonds
In case of a circular fingerprint, the minimum and maximum radius of the enumerated fragments can be specified. See Figure: Example of enumerated circular fragments with increasing radius
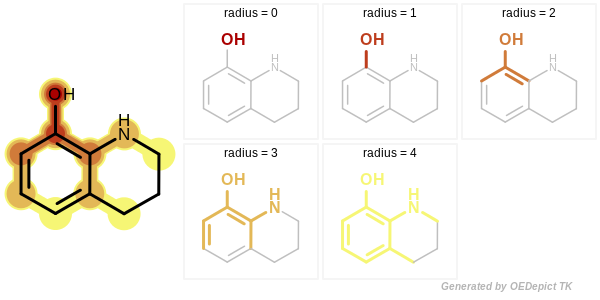
Example of enumerated circular fragments with increasing radius
For example, when generating a fingerprint of the molecule shown in
Figure: Example Molecule
with minimum and maximum length set to 0 and 3, respectively,
only paths listed in the first four rows in Table: Enumerated
Paths, are encoded into the fingerprint.
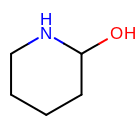
Example molecule
Path length (in bonds) |
Generated Unique Paths |
|---|---|
0 |
|
1 |
|
2 |
|
3 |
|
4 |
|
5 |
|
Figure: Example of enumerated paths depicts the six unique paths of length four that are generated for the example molecule. Each unique path is encoded only once without considering its frequency.
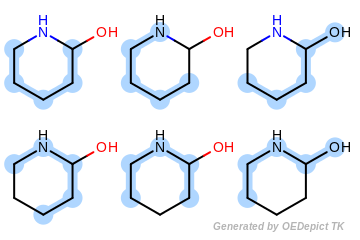
Example of enumerated paths
In the example shown in Listing 16, fingerprints
with various minimum and maximum path length are generated for pyrrole
and pyridine.
When enumerating only paths that are shorter than four bonds,
the fingerprints generated for the two molecules are identical.
Since the four bond-length pattern ccccc is present in pyridine
but not in pyrrole, the fingerprints become different, resulting in
a smaller Tanimoto similarity score.
Listing 16: Similarity calculation with various path lengths
public class FPPathLength
{
public static int Main(string[] args)
{
OEGraphMol molA = new OEGraphMol();
OEChem.OESmilesToMol(molA, "c1ccncc1");
OEGraphMol molB = new OEGraphMol();
OEChem.OESmilesToMol(molB, "c1cc[nH]c1");
PrintTanimoto(molA, molB, 0, 3);
PrintTanimoto(molA, molB, 1, 3);
PrintTanimoto(molA, molB, 0, 4);
PrintTanimoto(molA, molB, 0, 5);
return 0;
}
private static void PrintTanimoto(OEMolBase molA, OEMolBase molB, uint minb, uint maxb)
{
OEFingerPrint fpA = new OEFingerPrint();
OEFingerPrint fpB = new OEFingerPrint();
uint numbits = 2048;
uint atype = OEFPAtomType.DefaultAtom;
uint btype = OEFPBondType.DefaultBond;
OEGraphSim.OEMakePathFP(fpA, molA, numbits, minb, maxb, atype, btype);
OEGraphSim.OEMakePathFP(fpB, molB, numbits, minb, maxb, atype, btype);
Console.WriteLine("Tanimoto(A,B) = {0:0.000}", OEGraphSim.OETanimoto(fpA, fpB));
}
}
The output of Listing 16 is the following:
Tanimoto(A,B) = 1.000
Tanimoto(A,B) = 1.000
Tanimoto(A,B) = 0.950
Tanimoto(A,B) = 0.731
Fingerprint Size
The previous sections explain how the atom and bond typing and encoded fragment size can effect the similarity scores. Selecting an adequate fingerprint size is also very crucial. The number of unique circular, path or tree fragments present in molecular structures can be extremely large, therefore the generated fragments have to be hashed into the fixed-length fingerprint. This means that a bit in a fingerprint does not correspond to a unique pattern exclusively (as it does in structural key). Also a bit has no particular structural meaning, i.e., each bit represents the presence of a number of structural patterns.
The smaller the size of the fingerprints, the more dense they become, raising the probability of collisions. A collision occurs when different fragments are mapped to the same bit. This will inherently result in information loss and weaken the power to discriminate between structurally similar and dissimilar molecules. On the other hand, when the size of the fingerprints is too large they become very sparse, which will reduce information loss. However, the time spent to calculate similarity scores will increase.
The following table shows the number of unique paths generated for benzylpenicillin (depicted in Figure: Benzylpenicillin).
Note
The more atom and bond properties that are taken into account and the larger the size of paths to enumerate, the larger the size of the fingerprint has to be in order to encode the enumerated fragments without a significant number of bit collisions.
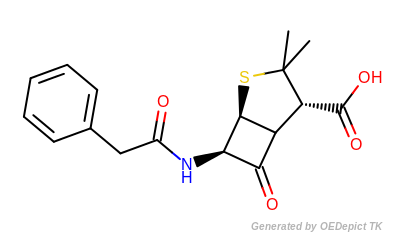
Benzylpenicillin
Atom/Bond typing |
path 0-3 |
path 0-5 |
path 0-7 |
|---|---|---|---|
56 |
149 |
297 |
|
111 |
265 |
453 |
|
126 |
297 |
499 |
|
147 |
362 |
617 |