Version 1.9.2
OEChem 1.9.2
New features
OEChem now supports the creation of InChI. OEChem produces the same InChI as the InChI library provided application for 99.77% of MDDR in the SDF format. The differences fall into two categories:
Corrupted molecules where the tools have to make arbitrary decisions about how to correctly interpret the data.
Bond stereo being perceived by the InChI application where bond stereo does not actually exist. OEChem does not assign bond stereo and generates what appears to be a more “correct” InChI for these cases.
The inclusion of InChI support added the following:
In addition, oemolostream will automatically write InChI when using the
.inchifile extension.OEPrepareSearchadded that perceives atom and bond properties of a molecule that are necessary to successfully execute a given substructure search. It is now highly recommended to run this function on a molecule before passing the molecule to any of OEChem’s graph matching algorithms.
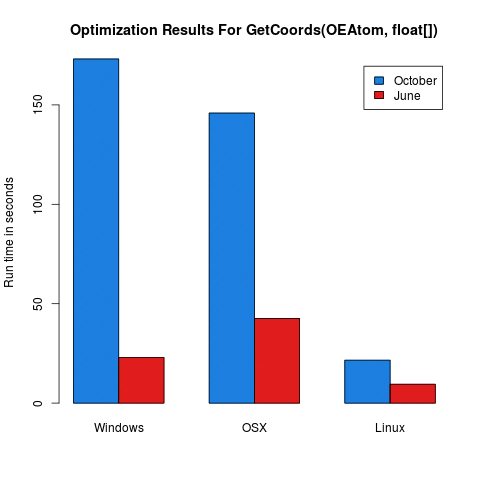
Several Java API points that pass primitive arrays of doubles or floats were optimized with good results. The following API points that take these primitive array types were optimized:
NewConf,SetCoords, andGetCoords.The following figures should be indicative of the performance increase in the 2013.June release: 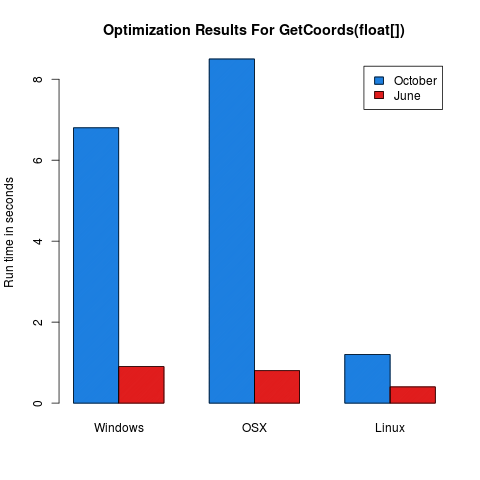
Changed the default input and output molecule file formats for molstreams and molthreads from
OEFormat.SMItoOEFormat.ISM.OEWriteMDLFilewrites MDL enhanced stereo groups.OEWriteMDLFilewill now automatically write a molecule in the V3000 file format if it has more than 999 atoms or if it has any MDL enhanced stereo group regardless of the given file format flavor.OESetMCSExhaustiveSearchTruncationLimitandOEGetMCSExhaustiveSearchTruncationLimitfunctions added that control how thoroughly the search is performed by theexhaustiveMCS search.Added the following standard SYBYL atom types:
OETriposType.CuOETriposType.Cooh,OETriposType.Croh,OETriposType.Crth,OETriposType.Fe,OETriposType.Mg,OETriposType.Mn,OETriposType.Mo,OETriposType.Se,OETriposType.Sn, andOETriposType.Zn. Atoms for which no SYBYL atom type exists are still internally handled with theOETriposType.Dutype, however when writing these atoms into aMOL2file their atomic symbol is written to the SYBYL atom type column instead of a string “Du”.Added
OEGetDimensionFromCoordsfunction that returns the dimension of the molecule from its coordinates.Residue information is now retained when reading files with MacroModel formats (.mmod, .mmd, and .dat).
Major bug fixes
It used to be possible to perform a substructure search for stereochemistry or hybridization on a molecule that did not yet have those properties perceived. This led to very subtle to detect problems where molecules would not match that were expected to match.
To combat this, the following OESubSearch methods now throw warnings in case some property that is necessary to successfully execute the substructure search has not been perceived yet:
These warnings can be suppressed by calling the
OEPrepareSearchfunction on the molecule first.Utilizing the
OEPerceived.AtomStereoandOEPerceived.BondStereoflag to mark whether the atom and bond stereo information is perceived in a molecule.It is now thread-safe to copy the same OEMol from multiple threads at the same time. This was caused by some OEMCMolBase methods not actually being const. The following methods were marked as const, but were not actually const, leading to subtle race conditions:
OEMCMolBase.GetMCMolTitlemethod added to provide a way to differentiate between a top-level OEMCMolBase title and a conformer title.OEMCSSearch.AddConstraintnow requires that the constraint is satisfied in all resulting matches. Previously, the documentation stated the following, “Constraints are considered satisfied in subgraphs which do not contain any constrained atoms or bonds in either the pattern or target molecules.” This is no longer the intended behavior.OEMCSSearch.AddConstraintnow returnsfalseif passed the same pattern atom or bond twice. The user must now useOEMCSSearch.ClearConstraintsin order to rebuild a set of constraints.
Minor bug fixes
OE3DToBondStereofunction returns false if the molecule dimension is 0, since no cis or trans bonds can be perceived if there are no coordinates.Fixed a problem when the max matches value of the OESubSearch was reset to the default value
1024every time the search was initialized by theOESubSearch.Initmethod.Adjusted the dimension of a molecule to 0 if it is read from either a
MDLor anSDFfile with no coordinates.OE3DToAtomStereofunction throws a warning and returnsfalseif any chiral atom of the molecule is not tetrahedral, i.e., if the atom stereo can not be determined from the 3D coordinates, usually because it is flat.The following functions and methods throw additional warnings in cases where either the
OE3DToAtomStereoor theOE3DToBondStereofunctions encounter any problems when assigning stereo from coordinates:OEPrepareSearchfunctionsOEWriteMoleculefunction when writing the following formats:OEFormat.MDL,OEFormat.SDFOEFormat.ISM,OEFormat.INCHIandOEFormat.INCHIKEYOEOmegaConfTest.CompareMolsmethod
OESubsetMolnow produces correct results when theRGroupflag wastrueand the molecule contained OEGroupBase objects.OEPerceiveChiralfunction considers hydrogens with different mass as being different i.e. it will recognize the C atom in a molecule “[3H]C(N)F” as a chiral atom, since it has four different neighbors: N, F, H and 3H.The first argument of the following methods is not const any longer:
OEOmegaConfTest.CompareMolsmethod
Improved the performance of the following methods:
OEOmegaConfTest.CompareMolsmethod
OEAtomBondSet.AddAtomandOEAtomBondSet.AddBondwill now return false when the trying to mix atoms from different molecules.OEAtomBondSet.HasAtomandOEAtomBondSet.HasBondadded for testing for membership.
The following functors are now properly wrapped in Python, Java, and C#:
The following functors are now deprecated:
OEChem::IsTrueAtom
OEChem::IsFalseAtom
OEChem::IsTrueBond
OEChem::IsFalseBond
OEChem::IsTrueConf
OEChem::IsFalseConf
Documentation fixes
The
OEAroModelnamespace is now accessible from the table of contents.
OESystem 1.9.2
New features
Added the following non-linear color gradients:
These classes along with the OELinearColorGradient class now derive from the OEColorGradientBase base class.
Minor bug fixes
OEPlatform 1.9.2
Major bug fixes
Calling OEPlatform::oeofstream::append after OEPlatform::oeofstream::open will no longer hang indefinitely once data is written to it.
Minor bug fixes
The java JVM can run out of memory when trying to free memory. If this is detected, the OpenEye toolkits will print the following message, “JNI::OutOfMemory – the JAVA heap may need to be increased – program terminating”. Please increase the size of the java heap using the
-Xmxoption.
OEBio 1.9.2
Minor internal improvements.
OEGrid 1.4.3
Minor bug fixes
OESkewGrid now properly initializes its base class OEGridAbstractBase during construction.
The
OEGridFileType.Asciiformat will now try harder to detect any incorrect file formatting issues and warn about them.
Documentation fixes
The
ASCIIWriterdocumentation example in the Grid Input and Output section was incorrect as it was writing out both grid indices and grid values.