Version 2.0.7
OEChem 2.0.7
New features
OEGetPathLengthfunction’s performance has been significantly improved.The APIs of the functions for accessing anisotropic B-factor parameters
OEGetAnisouandOESetAnisouhave been expanded for easier use in wrapped languages. The predicate OEHasAnisou and the container object OEAnisoUij have been added to support this functionality.The ability to request bond stereomark corrections to reduce the number of stereo-center correction warnings when reading MDL format files has been added. For more information, see
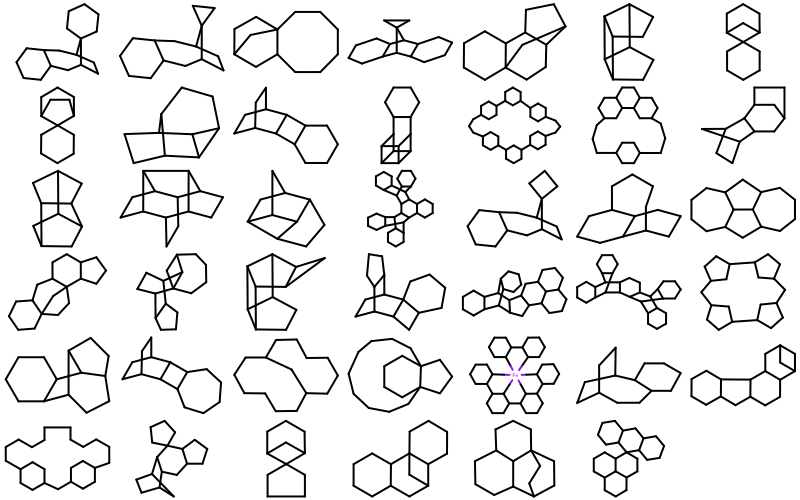
OEMDLStereoFromBondStereofunction,OEIFlavor.SDF.FixBondMarks,OEIFlavor.MDL.FixBondMarks, andOEIFlavor.RDF.FixBondMarksconstants.41 new ring templates have been added to the built-in ring dictionary.
Example of the new 2D ring templates Carbon Skeleton
Major bug fixes
OEMCSType.Approximatemethod for calculating the maximum-common-substructure of two molecules will no longer leak memory. This memory leak was introduced in the 2015.Oct release.The following functions and methods will no longer crash when used after
oemolithread.openoroemolothread.open, but will fail and returnfalse:
Minor bug fixes
The following problems have been fixed for reading V3000 MDL files:
OEReadMDLQueryFilefunction has been generalized to more reliably identify a V3000 header and dispatch to the correct reader.A parsing error for V3000 MDL files with an atom list has been fixed.
An issue with atom lists in V3000 format that resulted in the export of invalid atom symbols has been fixed. Atom lists will now export as “L” but will lose the original atom list information. Query features are not yet supported for molecule writing.
A new warning will be issued for any V3000 MDL files that contain SCSR template information. This information is currently ignored.
A new warning will be issued for any MDL files that contain Sgroup information or Rgroup definition. This information is currently ignored.
An issue that occurred when valid cis/trans bond stereo were being converted to
double eitherbonds in MDL reaction files has been corrected. This problem was introduced in the 2015.Oct release.The function to access anisotropic B-factor parameters
OEGetAnisounow sets all the parameters to0if no data is associated with the given atom, rather than leaving the parameters in an uninitialized state. The new predicate OEHasAnisou can be used to determine if an atom has these parameters.OEMCSSearch constructor no longer throws a warning when initialized with a molecule that has up-to-date ring perception information.
OEAssignAromaticFlagsfunction now invalidates the hybridization perception flag since different aromaticity models affect the recorded hybridization values.Atom stereochemistry is more thoroughly examined and may result in multiple marks to the stereocenter(s) and/or marks to ring bond(s) for
OEMDLStereoFromBondStereoactivity to reduce the number ofStereochemistry corrected ...warnings on re-read of the exported structure.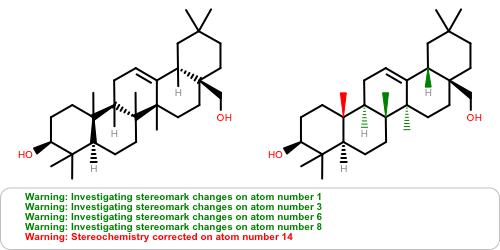
Documentation changes
Minor corrections to the
OEPerceivednamespace have been made.
OEBio 2.0.7
New features
OEPerceiveInteractionHintsfunction has been added to perceive possible ligand-protein interactions (contact, clash, hydrogen bond, salt-bridge, chelator, and aromatic ring stacking).The following classes and namespaces have been introduced to handle the new interaction types:
OEClashInteractionHint class and corresponding OEIsClashInteractionHint predicate
OESaltBridgeInteractionHint class and corresponding
OESaltBridgeInteractionHintTypenamespace and OEIsSaltBridgeInteractionHint predicateOEStackingInteractionHint class and corresponding
OEStackingInteractionHintTypenamespace and OEIsStackingInteractionHint predicate
OEGetActiveSiteInteractionHintTypesfunction has been added to return the interaction types perceived by the newOEPerceiveInteractionHintsfunction.OECrystalSymmetryParams class has been added, along with overloaded versions of
OEGetCrystalSymmetryandOESetCrystalSymmetryfunctions taking an OECrystalSymmetryParams object. This is the recommended API to use in the wrapped languages.
Minor bug fixes
OEMolComplexCategorizer.SetMaxPeptideResiduesmethod now properly sets the maximum number of residues that can be considered a peptide by OEMolComplexCategorizer.Lower-case chain IDs are now supported by
OESplitMolComplexand related functions.OERMSDnow checks whether the match stored in the given OESequenceAlignment object is valid prior to attempting to calculate RMSD.An uninitialized memory problem that occurred when
OEGetAlignmentfailed to perceive an acceptable alignment has been fixed.
Documentation changes
A link has been added from the OEResidue API documentation back to the Biopolymer Residues section to better clarify details about residue data.
A note has been added to the
OEResidue.IsHetAtomdocumentation concerning heterogen atoms being distinguished byHETATMrecords in a PDB file.Java and C# versions of examples Splitting a Macro-molecular Complex and Split a Macro-molecular Complex Into Fragments have been fixed.
Perceive and Print Protein-Ligand Interactions example has been updated to use the new OEInteractionHintContainer class and corresponding API.
OESystem 2.0.7
New features
OEGeom3DRotatetemplate math function can now be used with OEHalfFloat coordinate types.
OEPlatform 2.0.7
New features
OELIMITEDSTACKmacro define value can now be controlled from compiler flags like-DOELIMITEDSTACK=0. This allows projects to use the C++ header files when compiling OpenEye C++ code for use with OpenEye Java toolkits.For more information on compiling custom OpenEye C++ code for use by OpenEye Java toolkit projects, see https://github.com/oess/oejava_extension.