Depicting a Single Molecule
A program that converts a molecular structure into an image file.
Command Line Interface
A description of the command line interface can be obtained by executing the program with the –help argument.
prompt> python mol2img.py --help
will generate the following output:
Simple parameter list
image options :
-height : Height of output image
-width : Width of output image
input/output options :
-in : Input filename
-out : Output filename
-ringdict : User-defined 2D ring dictionary
molecule display options :
-aromstyle : Aromatic ring display style
-atomcolor : Atom coloring style
-atomlabelfontscale : Atom label font scale
-atomprop : Atom property display
-atomstereostyle : Atom stereo display style
-bondcolor : Bond coloring style
-bondprop : Bond property display
-bondstereostyle : Bond stereo display style
-hydrstyle : Hydrogen display style
-linewidth : Default bond line width
-protgroupdisp : Protective group display style
-scale : Scaling of the depicted molecule
-superdisp : Super atom display style
-titleloc : Location of the molecule title
prepare depiction options :
-clearcoords : Clear and regenerate 2D coordinates of molecule(s)
-orientation : Set the preferred orientation of 2D coordinates
-suppressH : Suppress explicit hydrogens of molecule(s)
Code
Download code
#!/usr/bin/env python
# (C) 2022 Cadence Design Systems, Inc. (Cadence)
# All rights reserved.
# TERMS FOR USE OF SAMPLE CODE The software below ("Sample Code") is
# provided to current licensees or subscribers of Cadence products or
# SaaS offerings (each a "Customer").
# Customer is hereby permitted to use, copy, and modify the Sample Code,
# subject to these terms. Cadence claims no rights to Customer's
# modifications. Modification of Sample Code is at Customer's sole and
# exclusive risk. Sample Code may require Customer to have a then
# current license or subscription to the applicable Cadence offering.
# THE SAMPLE CODE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND,
# EXPRESS OR IMPLIED. OPENEYE DISCLAIMS ALL WARRANTIES, INCLUDING, BUT
# NOT LIMITED TO, WARRANTIES OF MERCHANTABILITY, FITNESS FOR A
# PARTICULAR PURPOSE AND NONINFRINGEMENT. In no event shall Cadence be
# liable for any damages or liability in connection with the Sample Code
# or its use.
#############################################################################
# Converts a molecule structure into an image file.
# The output file format depends on its file extension.
#############################################################################
import sys
from openeye import oechem
from openeye import oedepict
def main(argv=[__name__]):
itf = oechem.OEInterface(InterfaceData)
oedepict.OEConfigureImageOptions(itf)
oedepict.OEConfigurePrepareDepictionOptions(itf)
oedepict.OEConfigure2DMolDisplayOptions(itf)
if not oechem.OEParseCommandLine(itf, argv):
oechem.OEThrow.Fatal("Unable to interpret command line!")
iname = itf.GetString("-in")
oname = itf.GetString("-out")
ext = oechem.OEGetFileExtension(oname)
if not oedepict.OEIsRegisteredImageFile(ext):
oechem.OEThrow.Fatal("Unknown image type!")
ifs = oechem.oemolistream()
if not ifs.open(iname):
oechem.OEThrow.Fatal("Cannot open input file!")
ofs = oechem.oeofstream()
if not ofs.open(oname):
oechem.OEThrow.Fatal("Cannot open output file!")
mol = oechem.OEGraphMol()
if not oechem.OEReadMolecule(ifs, mol):
oechem.OEThrow.Fatal("Cannot read input file!")
if itf.HasString("-ringdict"):
rdfname = itf.GetString("-ringdict")
if not oechem.OEInit2DRingDictionary(rdfname):
oechem.OEThrow.Warning("Cannot use user-defined ring dictionary!")
popts = oedepict.OEPrepareDepictionOptions()
oedepict.OESetupPrepareDepictionOptions(popts, itf)
oedepict.OEPrepareDepiction(mol, popts)
width, height = oedepict.OEGetImageWidth(itf), oedepict.OEGetImageHeight(itf)
dopts = oedepict.OE2DMolDisplayOptions(width, height, oedepict.OEScale_AutoScale)
oedepict.OESetup2DMolDisplayOptions(dopts, itf)
disp = oedepict.OE2DMolDisplay(mol, dopts)
oedepict.OERenderMolecule(ofs, ext, disp)
return 0
#############################################################################
# INTERFACE
#############################################################################
InterfaceData = """
!BRIEF [-in] <input> [-out] <output image> [-ringdict] <rd file>
!CATEGORY "input/output options :"
!PARAMETER -in
!ALIAS -i
!TYPE string
!REQUIRED true
!KEYLESS 1
!VISIBILITY simple
!BRIEF Input filename
!END
!PARAMETER -out
!ALIAS -o
!TYPE string
!REQUIRED true
!KEYLESS 2
!VISIBILITY simple
!BRIEF Output filename
!END
!PARAMETER -ringdict
!ALIAS -rd
!TYPE string
!REQUIRED false
!VISIBILITY simple
!BRIEF User-defined 2D ring dictionary
!DETAIL
2D ring dictionaries can be generated by the following OEChem examples:
C++ - createringdict.cpp
Python - createringdict.py
Java - CreateRingDict.java
C# - CreateRingDict.cs
!END
!END
"""
if __name__ == "__main__":
sys.exit(main(sys.argv))
See also
Molecule Depiction chapter
OEConfigureImageOptionsfunctionOEConfigurePrepareDepictionOptionsfunctionOEConfigure2DMolDisplayOptionsfunctionOEIsRegisteredImageFilefunctionOESetupPrepareDepictionOptionsfunctionOEPrepareDepictionfunctionOEGetImageWidthandOEGetImageHeightfunctionsOE2DMolDisplayOptions class
OESetup2DMolDisplayOptionsfunctionOE2DMolDisplay class
OERenderMoleculefunction
Examples
prompt> python mol2img.py -in nexium.ism -out image.png
will generate the image shown in Figure: Example of using the program with default parameters.
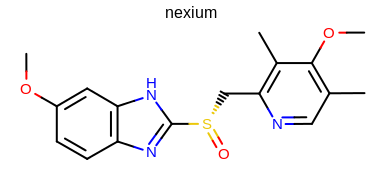
Example of using the program with default parameters
prompt> python mol2img.py -in nexium.ism -out image.png -atomcolor WhiteMonochrome
will generate the image shown in Figure: Example of using the program with the white monochrome atom coloring style.
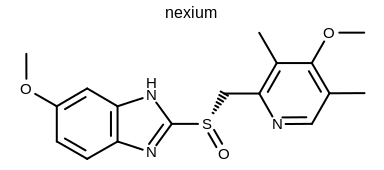
Example of using the program with the white monochrome atom coloring style
prompt> python mol2img.py -in nexium.ism -out image.png -aromstyle Circle
will generate the image shown in Figure: Example of using the program with aromaticity style ‘Circle’.
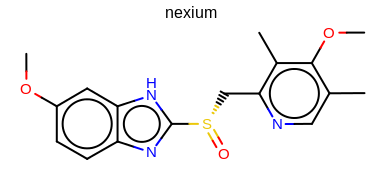
Example of using the program with aromaticity style ‘Circle’
prompt> python mol2img.py -in nexium.ism -out image.png -atomstereostyle "AtomStereo|CIPAtomStereo"
will generate the image shown in Figure: Example of the mol2img program with CIP atom stereo option.
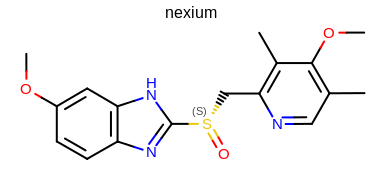
Example of using the program with CIP atom stereo option
prompt> python mol2img.py -in nexium.ism -out image.png -linewidth 4
will generate the image shown in Figure: Example of using the program with user-defined line width.
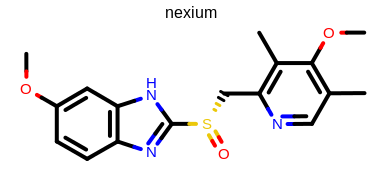
Example of using the program with user-defined line width
prompt> python mol2img.py -in nexium.ism -out image.png -superdisp All
will generate the image shown in Figure: Example of using the program with superatom style.
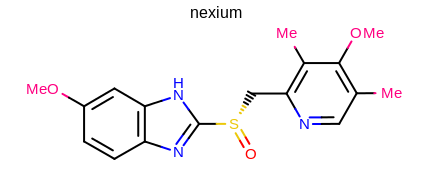
Example of using the program with superatom style
Note
The above program can generate 2D coordinates with user-defined 2D ring layouts when using the -ringdict parameter.
See also the 2D Coordinate Generation Examples section of the OEChem TK manual that shows examples about how to generate and utilize user-defined ring dictionaries.
See also
Molecule Depiction chapter