MDL Reaction Depiction
OEDepict TK also supports MDL reaction depiction.
The reaction being depicted has to be imported by calling the
OEReadMDLReactionQueryFile function.
The rest of the process is identical to depicting a molecule.
First the OEPrepareDepiction function has to be
called to generate the 2D atom coordinates, then the image can be
created by rendering the imported reaction (OERenderMolecule)
(See examples in Figure: Example of MDL reaction depiction)
Listing 1: Example of reaction depiction
if len(sys.argv) != 3:
oechem.OEThrow.Usage("%s <mdlreaction> <imagefile>" % sys.argv[0])
ifs = oechem.oemolistream(sys.argv[1])
qmol = oechem.OEGraphMol()
oechem.OEReadMDLReactionQueryFile(ifs, qmol)
oedepict.OEPrepareDepiction(qmol)
oedepict.OERenderMolecule(sys.argv[2], qmol)
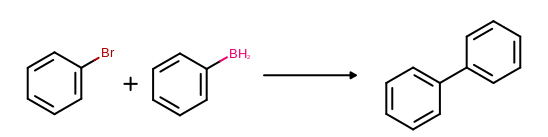
Example of MDL reaction depiction
See also
OEReadMDLReactionQueryFilefunction in the OEChem TK manualDepicting MDL Reaction example
The atom map information read from the reaction file can be depicted as atom properties by using the OEDisplayAtomMapIdx class. (See examples in Figure: Example of MDL reaction depiction with atom mapping)
Listing 2: Example of reaction depiction with map indices
if len(sys.argv) != 3:
oechem.OEThrow.Usage("%s <mdlreaction> <imagefile>" % sys.argv[0])
ifile = sys.argv[1]
ofile = sys.argv[2]
ifs = oechem.oemolistream(ifile)
qmol = oechem.OEGraphMol()
oechem.OEReadMDLReactionQueryFile(ifs, qmol)
oedepict.OEPrepareDepiction(qmol)
opts = oedepict.OE2DMolDisplayOptions()
opts.SetAtomPropertyFunctor(oedepict.OEDisplayAtomMapIdx())
disp = oedepict.OE2DMolDisplay(qmol, opts)
oedepict.OERenderMolecule(ofile, disp)
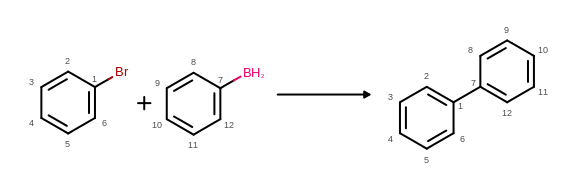
Example of MDL reaction depiction with atom mapping
See also
Listing 5example in the Molecule Depiction chapter