Molecule Alignment
The following three code examples demonstrate how to align molecules either based on:
maximum common substructure (
Listing 1),substructure matches (
Listing 2), ormolecular similarity (
Listing 3).
Molecule Alignment Based on MCS
The Figure: Example of depiction without alignment is generated by highlighting the maximum common substructure of two molecules (MCS), but keeping the original orientation of the compounds.
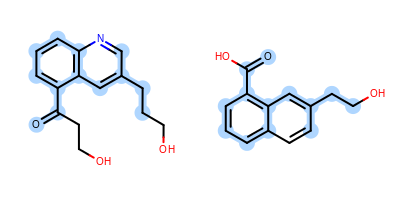
Example of depiction without alignment
The Listing 1 code example shows how to align these
two molecules by their maximum common substructures (i.e. by the
highlighted atoms and bonds in
Figure: Example of depiction without alignment).
First, two molecules are initialized from SMILES string and prepared for depiction.
A OEMCSSearch object is initialized with the reference 2D molecule onto which the fit molecule will be aligned.
An OEImageGrid object is constructed that allows to render the two molecules in a grid next to each other.
The fit molecule is then aligned to the reference by calling the
OEPrepareAlignedDepictionfunction that performs MCS search and then aligns the fit molecule to the reference based on the detected common substructure(s).An OE2DMolDisplayOptions object is initialized that stores the properties determine how the molecules are rendered. In order to render the molecules in equal size the smallest depiction scaling factor is calculated.
If the molecule alignment was successful (i.e
OEAlignmentResult.IsValidis true), then the correspondence between the two molecules stored in the OEAlignmentResult returned by theOEPrepareAlignedDepictionfunction. This OEAlignmentResult object can be used to highlight the matched common substructure by invoking theOEAddHighlightingfunction.After both molecules are rendered into the separate cells of the grid layout, the image is written into a
pngfile.
After initializing the two molecules and the OEMCSSearch object,
the OEMCSSearch.Match method is called to perform the
search and return an iterator over the identified maximum common
substructures.
The first match is then utilized to call the
OEPrepareAlignedDepiction function that aligns one molecule
to the other based on the given match.
After the alignment, the common atoms and bonds are highlighted by
invoking the OEAddHighlighting function.
The image created by Listing 1 is shown in
Figure: Example of depiction with alignment based on MCS.
Listing 1: Example of molecule alignment based on MCS
refmol = oechem.OEGraphMol()
oechem.OESmilesToMol(refmol, "c1cc(c2cc(cnc2c1)CCCO)C(=O)CCO")
oedepict.OEPrepareDepiction(refmol)
fitmol = oechem.OEGraphMol()
oechem.OESmilesToMol(fitmol, "c1cc2ccc(cc2c(c1)C(=O)O)CCO")
oedepict.OEPrepareDepiction(fitmol)
mcss = oechem.OEMCSSearch(oechem.OEMCSType_Approximate)
atomexpr = oechem.OEExprOpts_DefaultAtoms
bondexpr = oechem.OEExprOpts_DefaultBonds
mcss.Init(refmol, atomexpr, bondexpr)
mcss.SetMCSFunc(oechem.OEMCSMaxBondsCompleteCycles())
alignres = oedepict.OEPrepareAlignedDepiction(fitmol, mcss)
image = oedepict.OEImage(400, 200)
rows, cols = 1, 2
grid = oedepict.OEImageGrid(image, rows, cols)
opts = oedepict.OE2DMolDisplayOptions(grid.GetCellWidth(), grid.GetCellHeight(),
oedepict.OEScale_AutoScale)
opts.SetTitleLocation(oedepict.OETitleLocation_Hidden)
refscale = oedepict.OEGetMoleculeScale(refmol, opts)
fitscale = oedepict.OEGetMoleculeScale(fitmol, opts)
opts.SetScale(min(refscale, fitscale))
refdisp = oedepict.OE2DMolDisplay(mcss.GetPattern(), opts)
fitdisp = oedepict.OE2DMolDisplay(fitmol, opts)
if alignres.IsValid():
refabset = oechem.OEAtomBondSet(alignres.GetPatternAtoms(), alignres.GetPatternBonds())
oedepict.OEAddHighlighting(refdisp, oechem.OEBlueTint,
oedepict.OEHighlightStyle_BallAndStick, refabset)
fitabset = oechem.OEAtomBondSet(alignres.GetTargetAtoms(), alignres.GetTargetBonds())
oedepict.OEAddHighlighting(fitdisp, oechem.OEBlueTint,
oedepict.OEHighlightStyle_BallAndStick, fitabset)
refcell = grid.GetCell(1, 1)
oedepict.OERenderMolecule(refcell, refdisp)
fitcell = grid.GetCell(1, 2)
oedepict.OERenderMolecule(fitcell, fitdisp)
oedepict.OEWriteImage("MCSAlign.png", image)
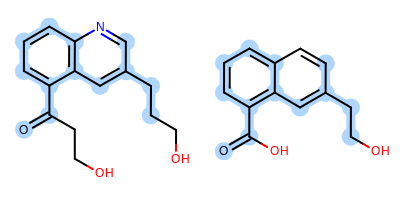
Example of depiction with alignment based on maximum common substructure
See also
OEMCSSearch class in the OEChem TK manual
OEImageGrid class
OEAddHighlightingfunctionOERenderMoleculefunctionOEWriteImagefunctionAligning Molecule Based on MCS example
Molecule Alignment Based on Substructure Search
The Figure: Example of depiction without alignment is generated by highlighting the “O=C(O)C(N)” substructures in four amino acids, but keeping the original orientation of the compounds.
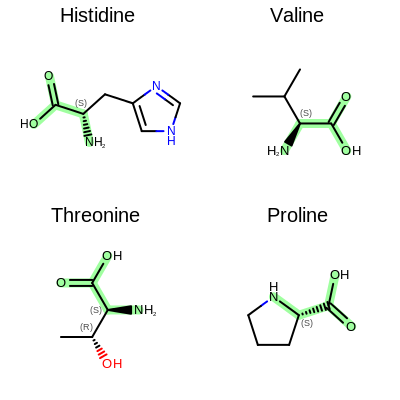
Example of depiction without alignment
The Listing 2 code example shows how to align these
molecules by their matched substructures (i.e. by the highlighted atoms and bonds
in Figure: Example of depiction without alignment).
First, the OESubSearch object is initialized with the reference 2D molecule onto which all amino acid structures will be aligned.
Each amino acids structure is initialized from a SMILES string and prepared for depiction.
An OEImageGrid object is constructed that allows to render the molecules into a grid layout.
An OE2DMolDisplayOptions object is initialized that stores the properties determine how the molecules are rendered. In order to render the molecules in equal size the smallest depiction scaling factor is calculated.
Looping over the amino acids:
Each molecule is aligned to the reference molecule by calling the
OEPrepareAlignedDepictionfunction that performs the alignment based on the detected substructure search match(es).If the molecule alignment was successful (i.e
OEAlignmentResult.IsValidis true), then the correspondence between the two molecules stored in the OEAlignmentResult returned by theOEPrepareAlignedDepictionfunction. This OEAlignmentResult object can be used to highlight the matched substructure by invoking theOEAddHighlightingfunction.The aligned (and highlighted) molecule is then rendered to a next cell of the OEImageGrid object.
Finally, the image is written into a
pngfile.
The image created by Listing 2 is shown in
Figure: Example of depiction with alignment based on substructure search.
Listing 2: Example of molecule alignment based on substructure search
refmol = oechem.OEGraphMol()
oechem.OESmilesToMol(refmol, "O=C(O)C(N)")
oedepict.OEPrepareDepiction(refmol)
ss = oechem.OESubSearch(refmol, oechem.OEExprOpts_DefaultAtoms, oechem.OEExprOpts_DefaultBonds)
aminosmiles = ["O=C(O)[C@@H](N)Cc1c[nH]cn1 Histidine",
"CC(C)[C@@H](C(=O)O)N Valine",
"C[C@H]([C@@H](C(=O)O)N)O Threonine",
"C1C[C@H](NC1)C(=O)O Proline"]
aminoacids = []
for smi in aminosmiles:
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, smi)
oedepict.OEPrepareDepiction(mol)
aminoacids.append(oechem.OEGraphMol(mol))
image = oedepict.OEImage(400, 400)
rows, cols = 2, 2
grid = oedepict.OEImageGrid(image, rows, cols)
opts = oedepict.OE2DMolDisplayOptions(grid.GetCellWidth(),
grid.GetCellHeight(), oedepict.OEScale_AutoScale)
opts.SetAtomStereoStyle(oedepict.OEAtomStereoStyle_Display_All)
minscale = float("inf")
for amino in aminoacids:
minscale = min(minscale, oedepict.OEGetMoleculeScale(amino, opts))
opts.SetScale(minscale)
for amino, cell in zip(aminoacids, grid.GetCells()):
alignres = oedepict.OEPrepareAlignedDepiction(amino, ss)
disp = oedepict.OE2DMolDisplay(amino, opts)
if alignres.IsValid():
oedepict.OEAddHighlighting(disp, oechem.OELightGreen,
oedepict.OEHighlightStyle_Stick, alignres)
oedepict.OERenderMolecule(cell, disp)
oedepict.OEWriteImage("SubSearchAlign.png", image)
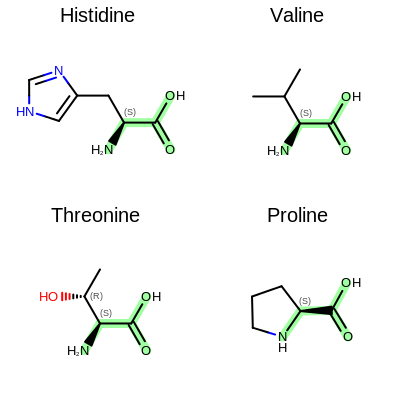
Example of depiction with alignment based on substructure match
See also
OESubSearch class in the OEChem TK manual
OEImageGrid class
OEAddHighlightingfunctionOERenderMoleculefunctionOEWriteImagefunction
Molecule Alignment Based on Molecular Similarity
The Figure: Example of depiction without alignment is generated by keeping the original orientation of the compounds.
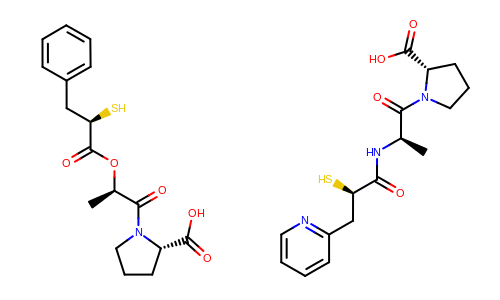
Example of depiction without alignment
The Listing 3 code example shows how to align these
molecules based on their molecular similarity.
First, two molecules are initialized from SMILES string and prepared for depiction.
An OEImageGrid object is constructed that allows to render the two molecules in a grid next to each other.
The fit molecule is then aligned to the reference. The
OEGetFPOverlapfunction is utilized to return all common fragments found between two molecules based on a given fingerprint type. These common fragments reveal the similar parts of the two molecules being compared that are used by theOEPrepareMultiAlignedDepictionfunction to find the best alignment between the molecules. See more fingerprint types in the Fingerprint Types chapter of the GraphSim TK manual.An OE2DMolDisplayOptions object is initialized that stores the properties determine how the molecules are rendered. In order to render the molecules in equal size the smallest depiction scaling factor is calculated.
Then both molecules are rendered into the separate cells of the grid layout, the image is written into a
pngfile.
The image created by Listing 3 is shown in
Figure: Example of depiction with alignment based on molecular similarity.
Note
GraphSim TK license in not required to run the Listing 3 example.
Listing 3: Example of molecule alignment based on molecular similarity
refmol = oechem.OEGraphMol()
oechem.OESmilesToMol(refmol, "C[C@H](C(=O)N1CCC[C@H]1C(=O)O)OC(=O)[C@@H](Cc2ccccc2)S")
oedepict.OEPrepareDepiction(refmol)
fitmol = oechem.OEGraphMol()
oechem.OESmilesToMol(fitmol, "C[C@H](C(=O)N1CCC[C@H]1C(=O)O)NC(=O)[C@@H](Cc2ccccn2)S")
oedepict.OEPrepareDepiction(fitmol)
image = oedepict.OEImage(500, 300)
rows, cols = 1, 2
grid = oedepict.OEImageGrid(image, rows, cols)
overlaps = oegraphsim.OEGetFPOverlap(refmol, fitmol,
oegraphsim.OEGetFPType(oegraphsim.OEFPType_Tree))
oedepict.OEPrepareMultiAlignedDepiction(fitmol, refmol, overlaps)
opts = oedepict.OE2DMolDisplayOptions(grid.GetCellWidth(), grid.GetCellHeight(),
oedepict.OEScale_AutoScale)
opts.SetTitleLocation(oedepict.OETitleLocation_Hidden)
refscale = oedepict.OEGetMoleculeScale(refmol, opts)
fitscale = oedepict.OEGetMoleculeScale(fitmol, opts)
opts.SetScale(min(refscale, fitscale))
refdisp = oedepict.OE2DMolDisplay(refmol, opts)
fitdisp = oedepict.OE2DMolDisplay(fitmol, opts)
refcell = grid.GetCell(1, 1)
oedepict.OERenderMolecule(refcell, refdisp)
fitcell = grid.GetCell(1, 2)
oedepict.OERenderMolecule(fitcell, fitdisp)
oedepict.OEWriteImage("FPAlign.png", image)
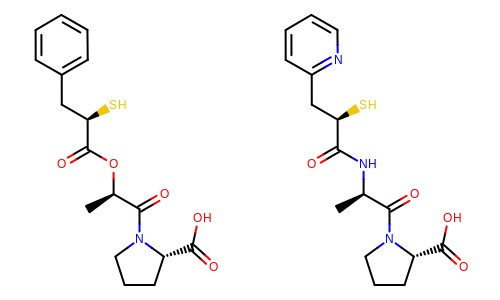
Example of depiction with alignment based on molecule similarity
See also
OEImageGrid class
OERenderMoleculefunctionOEWriteImagefunction
See also
The Python script that visualizes molecule similarity based on fingerprints can be downloaded from the OpenEye Python Cookbook