Matched Pair analysis and transformations
A program that loads a previously generated Matched Pair index, then reads an input set of structures and outputs transformed structures based on the matched pairs discovered during indexing.
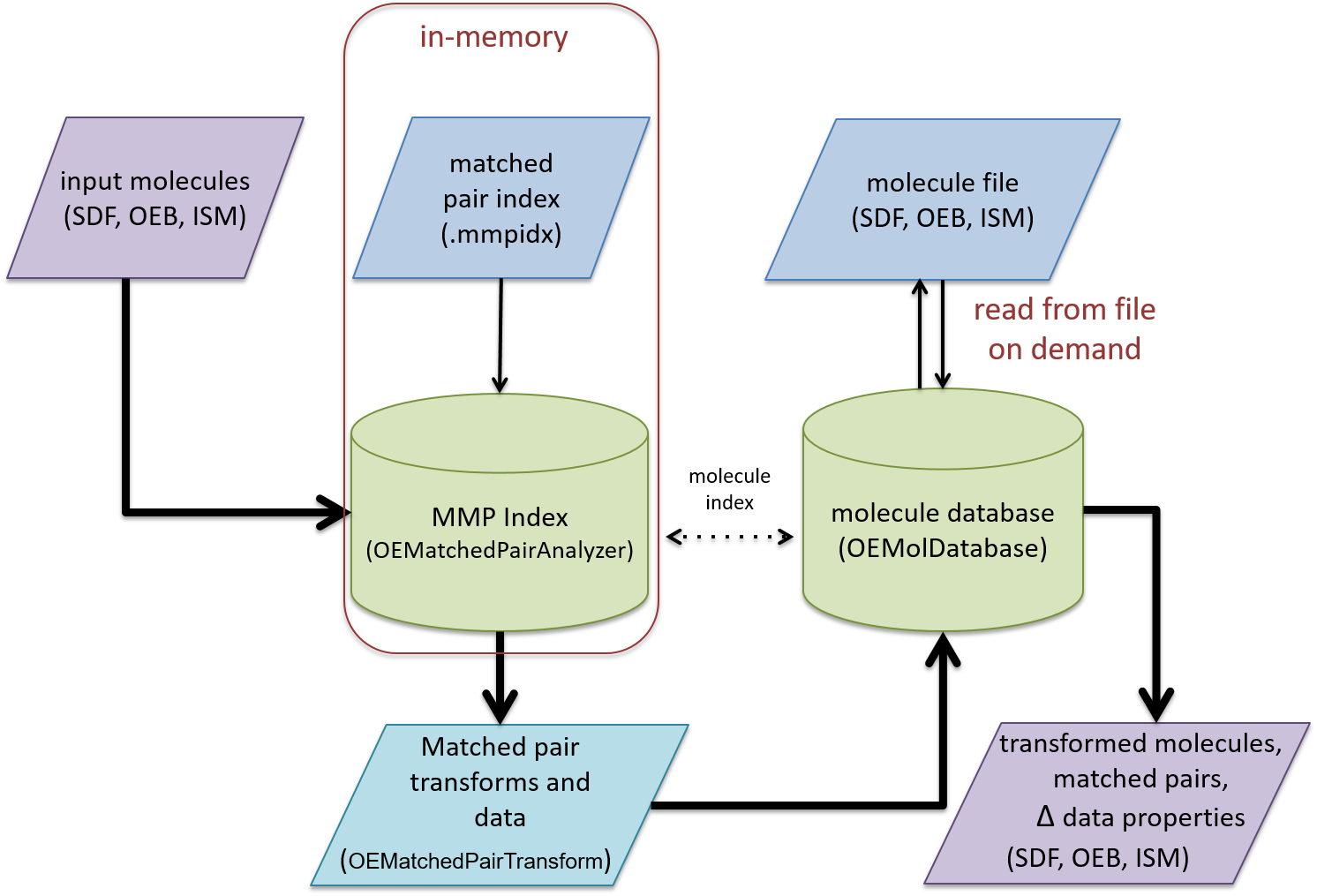
Schematic representation of the Matched Pair Analysis process
See also
OEMatchedPairAnalyzer class
OEMatchedPairApplyTransformsfunction
Command Line Interface
prompt> MatchedPairTransform.py -mmpidx index.mmpidx -input input.sdf -output output.sdf
Code
Download code
#!/usr/bin/env python
# (C) 2022 Cadence Design Systems, Inc. (Cadence)
# All rights reserved.
# TERMS FOR USE OF SAMPLE CODE The software below ("Sample Code") is
# provided to current licensees or subscribers of Cadence products or
# SaaS offerings (each a "Customer").
# Customer is hereby permitted to use, copy, and modify the Sample Code,
# subject to these terms. Cadence claims no rights to Customer's
# modifications. Modification of Sample Code is at Customer's sole and
# exclusive risk. Sample Code may require Customer to have a then
# current license or subscription to the applicable Cadence offering.
# THE SAMPLE CODE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND,
# EXPRESS OR IMPLIED. OPENEYE DISCLAIMS ALL WARRANTIES, INCLUDING, BUT
# NOT LIMITED TO, WARRANTIES OF MERCHANTABILITY, FITNESS FOR A
# PARTICULAR PURPOSE AND NONINFRINGEMENT. In no event shall Cadence be
# liable for any damages or liability in connection with the Sample Code
# or its use.
#############################################################################
# Utility to load a previously generated MMP index
# and use the transformations discovered to alter a second set of structures
# ---------------------------------------------------------------------------
# MatchedPairTransform.py mmp_index input_mols output_mols
#
# mmp_index: filename of matched pair index
# input_mols: filename of molecules to transform based on analysis
# output_mols: filename to collect transformed molecules
#############################################################################
from openeye import oechem
from openeye import oemedchem
import sys
def MMPTransform(itf):
# input structure(s) to process
ifsmols = oechem.oemolistream()
if not ifsmols.open(itf.GetString("-input")):
oechem.OEThrow.Fatal("Unable to open %s for reading" %
itf.GetString("-input"))
# check MMP index
mmpimport = itf.GetString("-mmpindex")
if not oemedchem.OEIsMatchedPairAnalyzerFileType(mmpimport):
oechem.OEThrow.Fatal('Not a valid matched pair index input file, {}'.format(mmpimport))
# load MMP index
mmp = oemedchem.OEMatchedPairAnalyzer()
if not oemedchem.OEReadMatchedPairAnalyzer(mmpimport, mmp):
oechem.OEThrow.Fatal("Unable to load index {}".format(mmpimport))
if not mmp.NumMols():
oechem.OEThrow.Fatal('No records in loaded MMP index file: {}'.format(mmpimport))
if not mmp.NumMatchedPairs():
oechem.OEThrow.Fatal('No matched pairs found in MMP index file, ' +
'use -fragGe,-fragLe options to extend indexing range')
# output (transformed) structure(s)
ofs = oechem.oemolostream()
if not ofs.open(itf.GetString("-output")):
oechem.OEThrow.Fatal("Unable to open %s for writing" %
itf.GetString("-output"))
# request a specific context for the transform activity, here 0-bonds
chemctxt = oemedchem.OEMatchedPairContext_Bond0
askcontext = itf.GetString("-context")[:1]
if askcontext == '0':
chemctxt = oemedchem.OEMatchedPairContext_Bond0
elif askcontext == '1':
chemctxt = oemedchem.OEMatchedPairContext_Bond1
elif askcontext == '2':
chemctxt = oemedchem.OEMatchedPairContext_Bond2
elif askcontext == '3':
chemctxt = oemedchem.OEMatchedPairContext_Bond3
elif askcontext == 'a' or askcontext == 'A':
chemctxt = oemedchem.OEMatchedPairContext_AllBonds
else:
oechem.OEThrow.Fatal("Invalid context specified: " +
askcontext + ", only 0|1|2|3|A allowed")
verbose = itf.GetBool("-verbose")
# return some status information
if verbose:
oechem.OEThrow.Info("{}: molecules: {:d}, matched pairs: {:,d}"
.format(mmpimport,
mmp.NumMols(),
mmp.NumMatchedPairs()))
minpairs = itf.GetInt("-minpairs")
if minpairs > 1 and verbose:
oechem.OEThrow.Info('Requiring at least %d matched pairs to apply transformations'
% minpairs)
errs = None
if itf.GetBool("-nowarnings"):
errs = oechem.oeosstream()
oechem.OEThrow.SetOutputStream(errs)
orec = 0
ocnt = 0
for mol in ifsmols.GetOEGraphMols():
orec += 1
iter = oemedchem.OEMatchedPairApplyTransforms(mol, mmp, chemctxt, minpairs)
if not iter.IsValid():
if verbose:
# as minpairs increases, fewer transformed mols are generated - output if requested
name = mol.GetTitle()
if not mol.GetTitle():
name = 'Record ' + str(orec)
oechem.OEThrow.Info("%s did not produce any output" % name)
continue
if errs is not None:
errs.clear()
for outmol in iter:
ocnt += 1
oechem.OEWriteMolecule(ofs, outmol)
if errs is not None:
errs.clear()
if not orec:
oechem.OEThrow.Fatal('No records in input structure file to transform')
if not ocnt:
oechem.OEThrow.Warning('No transformed structures generated')
print("Input molecules={} Output molecules={}".format(orec, ocnt))
return 0
############################################################
InterfaceData = """
# matchedpairtransform interface file
!CATEGORY MatchedPairTransform
!CATEGORY I/O
!PARAMETER -mmpindex 1
!TYPE string
!REQUIRED true
!BRIEF Input filename of serialized matched pair index to load
!END
!PARAMETER -input 2
!ALIAS -i
!ALIAS -in
!TYPE string
!REQUIRED true
!BRIEF Input filename of structures to process using matched pairs from -mmpindex
!END
!PARAMETER -output 3
!ALIAS -o
!ALIAS -out
!TYPE string
!REQUIRED true
!BRIEF Output filename
!END
!END
!CATEGORY options
!PARAMETER -context 1
!ALIAS -c
!TYPE string
!DEFAULT 0
!BRIEF chemistry context to use for the transformation [0|1|2|3|A]
!END
!PARAMETER -minpairs 2
!TYPE int
!DEFAULT 0
!BRIEF require at least -minpairs to apply the transformations (default: all)
!END
!PARAMETER -verbose 3
!TYPE bool
!DEFAULT 0
!BRIEF generate verbose output
!END
!PARAMETER -nowarnings 4
!TYPE bool
!DEFAULT 1
!BRIEF suppress warning messages from applying transformations
!END
!END
!END
"""
def main(argv=[__name__]):
itf = oechem.OEInterface(InterfaceData)
if not oechem.OEParseCommandLine(itf, argv):
oechem.OEThrow.Fatal("Unable to interpret command line!")
MMPTransform(itf)
if __name__ == "__main__":
sys.exit(main(sys.argv))