Version 2.0.6
OEChem 2.0.6
New features
The performance of the SDF V3000 format file parsing has been significantly improved and is now approximately 50% faster. The speed of importing an SDF file in V3000 format is now comparable to V2000. See table Performance improvement of importing SDF V3000 file format below that shows the improvements.
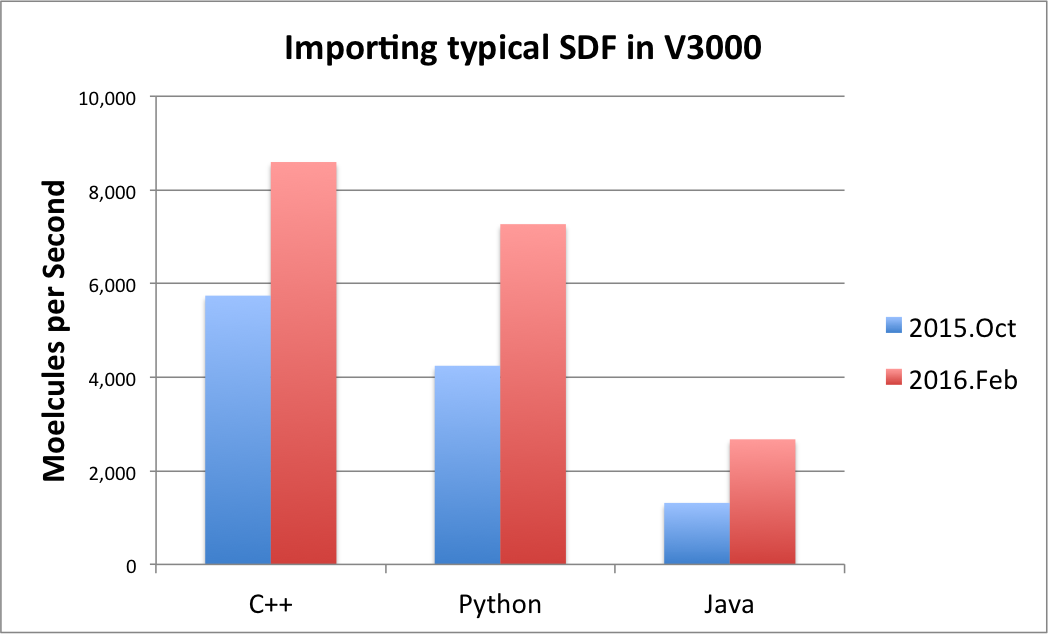
|
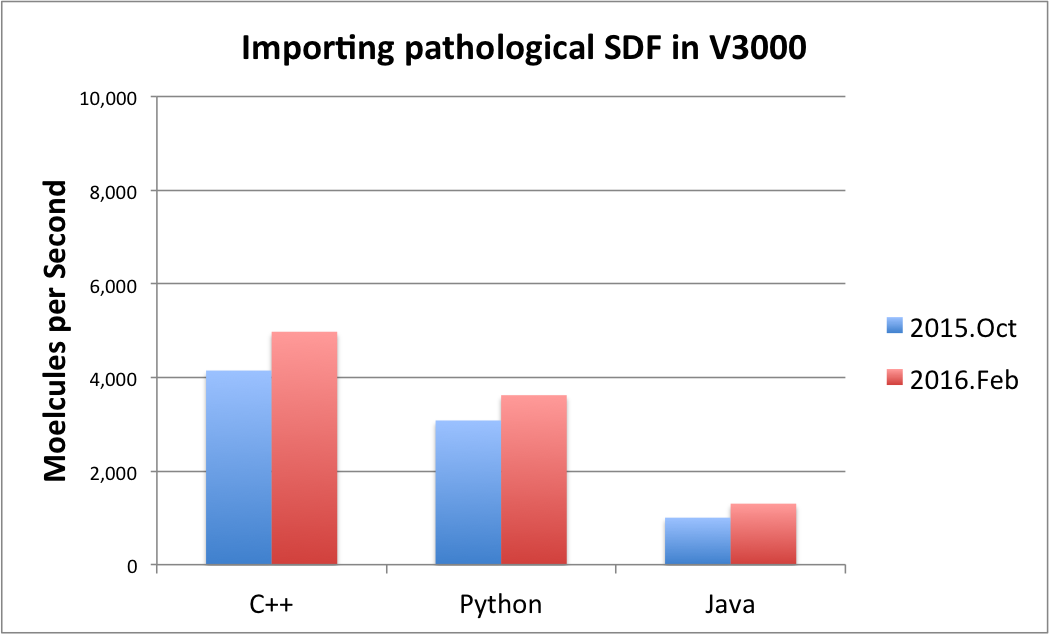
|
Note
There are some pathological cases when only a slight improvement has been achieved. From the parsing viewpoint, a pathological or atypical SDF V3000 file contains an excessive number of non-default property values, redundantly specified atom or bond properties, and/or atom coordinates in scientific notation which requires a more general but slower parsing activity. For typical SDF V3000 files, a significant improvement was seen, but generally the magnitude of the improvement is largely input data dependent.
OEWriteMolToStringandOEReadMolFromStringoverload functions have been added to allow specification of the file format using the the OEFormat namespace.
OEMolBase::Clearperformance has been improved whenever the molecule is already empty.The following flavors have been added to the
OEOFlavor::MOL2namespace:OEOFlavor::MOL2::Forcefield,OEOFlavor::MOL2::ChargePrecision, andOEOFlavor::MOL2::GeneralFFFormat. These flavors allow writing nonstandard variants of Tripos MOL2 files targeted at the general force field community.
Major bug fixes
OEMMFF94PartialChargesfunction no longer crashes with incorrect atom types.OEUniMolecularRxnfunction no longer crashes when bonds to mapped explicit hydrogens are deleted.
Minor bug fixes
OESortConfsByTagfunction can now sort conformations by generic data with double type.OE2DRingDictionary::AddRingsmethod now allows the addition of ring templates with extremely high average bond length. These ring templates are normalized before inserting them into the ring dictionary.OEChem TK’s MDL V2000 and V3000 readers have been improved to handle nonstandard or incorrect MDL or SDF files.
OEReadMoleculenow warns about invalid bond stereo marks on non-single bond types and will ignore them. Additionally, a common error in CTfile format files is the presence of a “wedge either” bond on a double bond. This latter error is now automatically changed to the assumed (non-wedge) “double either” bond mark and a warning is generated.OEReadMoleculenow attempts to read a variant of the SDFile format that contains blank line(s) before the start of the SD data appendices. Although this is a deviation from the CTfile format, this format has been known to occur in the wild. This change now impacts the use of concatenated MOL files when the structures have blank molecule titles. In general, concatenated MOL files are a much less preferred strategy for multiple record structure input and should be avoided. It is highly recommended to always use SDF files for multi-record input since an explicit record delimiter is always present.When
OEReadMoleculeencounters a connection table format error for SDF format files, it now advances to the next record delimiter. Previously, it would have attempted to reset and reread at an arbitrary point in the corrupted file, possibly generating additional warnings.OEReadMoleculeand low level MDL format readers are now more tolerant for V3000 format files that contain arbitrary collection types. Previously, only stereo collections and highlight collections were allowed. Now a message about unknown types generates the warningSkipping unknown collection type, XXX/YYY, withXXX/YYYindicating the specific collection type that was ignored. Unknown collection information is not persisted to any output format types: it is well and truly skipped!A warning is now thrown when multiple rgroup label sites (e.g., R1R2) are encountered, indicating that this type of representation is not yet supported.
When reading V3000 format containing pseudo-atoms (i.e., atoms not in the internal OpenEye element list), the atom symbol information is no longer lost but can be retrieved from
OEAtomBase::GetNameand is now the same as V2000 file format handling.
When an
OEMolBaseType::OEMiniMolmolecule implementation is instantiated from another OEMolBase instance that contains one or more atoms, the dimension code is also copied so thatOEMolBase::GetDimensionmatches the dimension setting from the original OEMolBase instance.OEReadMoleculeis now more tolerant for SKC format files containing explicit string tags of 0-length.OEAssignHybridizationnow ignores transition metals, lanthanides, and actinides and sets their hybridization toOEHybridization::Unknown. As a result, these atoms are no longer inadvertently considered to be potential tetrahedral stereocenters.OEMolDatabase cannot support file formats without explicit record delimiters, so files such as MOL, MDL, and RXN cannot be supported. A properly formatted SDF file is the preferred input to initialize the OEMolDatabase class.
OEMolDatabase now fails early and refuses to parse the junk data when a file changes underneath an OEMolDatabase. This can happen when an NFS client changes a file that is already open on another NFS directory, invalidating the NFS client that is using OEMolDatabase.
OESweepRotorCompressHydrogensno longer returnsfalsewhen the molecule passed in does not contain any hydrogens. However, it returnsfalseif the molecule contains any deleted atoms, as it is then likely that the rotor compression data is already corrupted.OEGeom3DMatrixInvertfunction has been fixed.A bug that allowed the
!DEFAULTvalue of a parameter in the configuration file to be set to a value that is illegal for that parameter has been fixed. A warning is now thrown when an illegal value is set.A bug that caused PDB Data records, such as
REMARKandSSBON, to be clipped at 72 characters instead of 80 characters has been fixed.
C++-specific changes
A const OEGraphMol, OEMol, or OEQMol no longer results in a compilation error when using the following generic data getters that should have been previously marked
constin the header file:GetBoolData,GetIntData,GetFloatData,GetDoubleData, andGetStringData.OEPRECompressthat outputs an OEMol has been deprecated in favor of theOEPrepareFastROCSMolfunction. This function will be removed in a future release.Note
OEPRECompressthat alters the binary IO handlers on an oemolstreambase is still supported and is the appropriate way to turn on PRE-compressedOEB.
Python-specific changes
Passing
Noneto the oemolistream and oemolostream constructors no longer results in a crash.
Documentation changes
OEDeleteEverythingExceptTheFirstLargestComponentfunction is now documented.
OEBio 2.0.6
New features
OESplitMolComplexOptions::SetSplitCovalentCofactorsandOESplitMolComplexOptions::GetSplitCovalentCofactorsmethods have been added to control the splitting of covalent cofactors from a macromolecular complex. The new constantOESplitMolComplexSetup::CovCofactorcontrols whetherOEConfigureSplitMolComplexOptionssets up command-line parsing for this option.OEMolComplexCategorizer can now recognize a multi-residue OEAtomBondSet as a covalently attached ligand or cofactor.
AminoAcid, a new residue database category, has been introduced. It consists of standard amino acids and common variants such as seleno-methionine. Previously, these had been listed in the categoryCofactor.OESplitMolComplexOptions::SetWarnNoLigandandOESplitMolComplexOptions::GetWarnNoLigandmethods have been added to control whether the molecular complex splitting functions generate a verbose message warning whenever a ligand is not identified.OEClearMolComplexSDDatafunction has been added to remove SD tags generated byOEGetMolComplexComponents.OEGetAlignmentshas been added to deal with multiple chains in each structure. The method returns an iterator of alignments, one for each pairwise chain alignment.OEGetAlignmentnow returns the highest scored alignment fromOEGetAlignments.OEGetSimpleAlignmentis a replacement forOEGetAlignment, which only looks at the first chain in each sequence.OEWriteAlignmenthas gained a third parameter to allow varying the width of the output.The following methods have been added to the OESequenceAlignment class:
Minor bug fixes
Titles generated by functions
OESplitMolComplexandOEGetMolComplexComponentsno longer contain single quotes or blank characters.Ongoing maintenance has been performed in the OEResidueCategoryData database used by OESplitMolComplexOptions. Residues have been removed from the
PolymerandMisclists.
Documentation changes
The examples have been updated to perceive residues when this is not performed by the default molecule reader activity (for example, in the case of
.mol2). With this update, examples that had previously been transforming input hydrogen names from a PDB file to the new nomenclature (closecontacts,makealpha,subsetres, andswapaieres) now retain the input hydrogen names.
OESystem 2.0.6
New features
OEThreadedDots has been added to provide a thread-safe way to output progress bar dots to the terminal when the work is being updated from multiple threads simultaneously.
OEPlatform 2.0.6
Minor internal improvements have been made.
OEGrid 1.5.3
Major bug fixes
OEMakeGridFromCenterAndExtentsnow propagates errors with the grid construction.OEReadGridcan now read a gzipped OESystem::OEFizzGrid.