Version 2.0.5
OEChem 2.0.5
New features
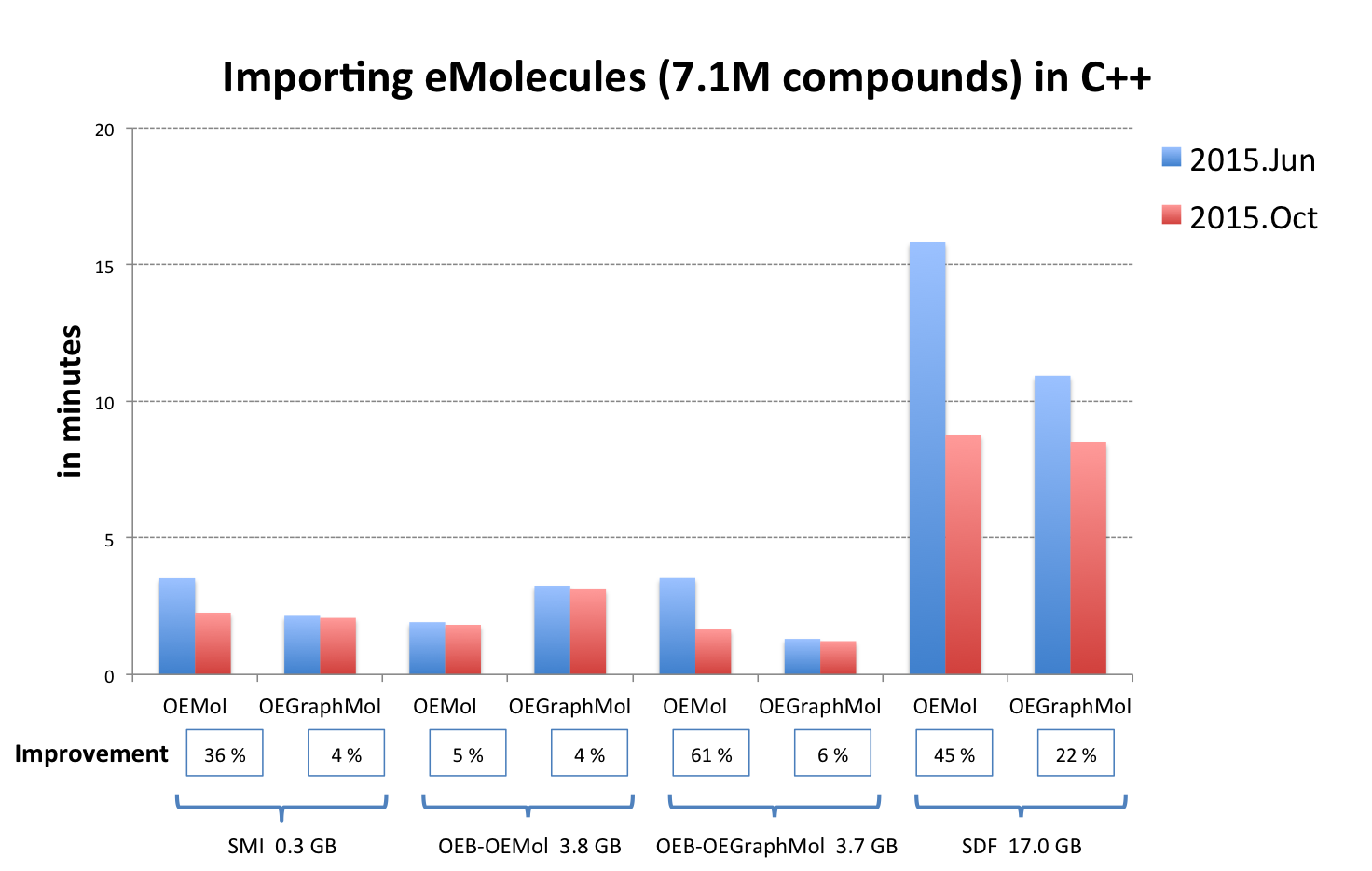
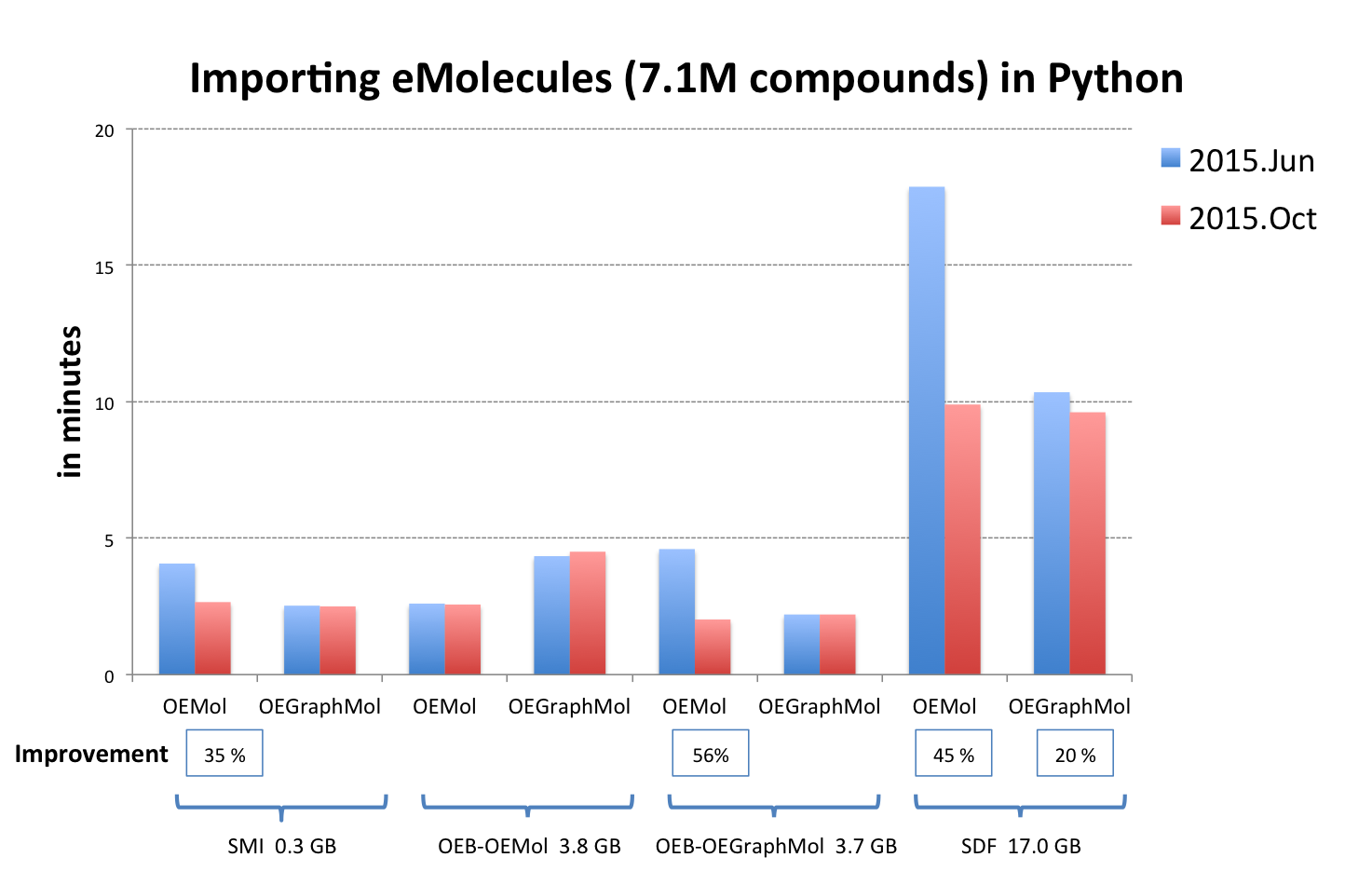
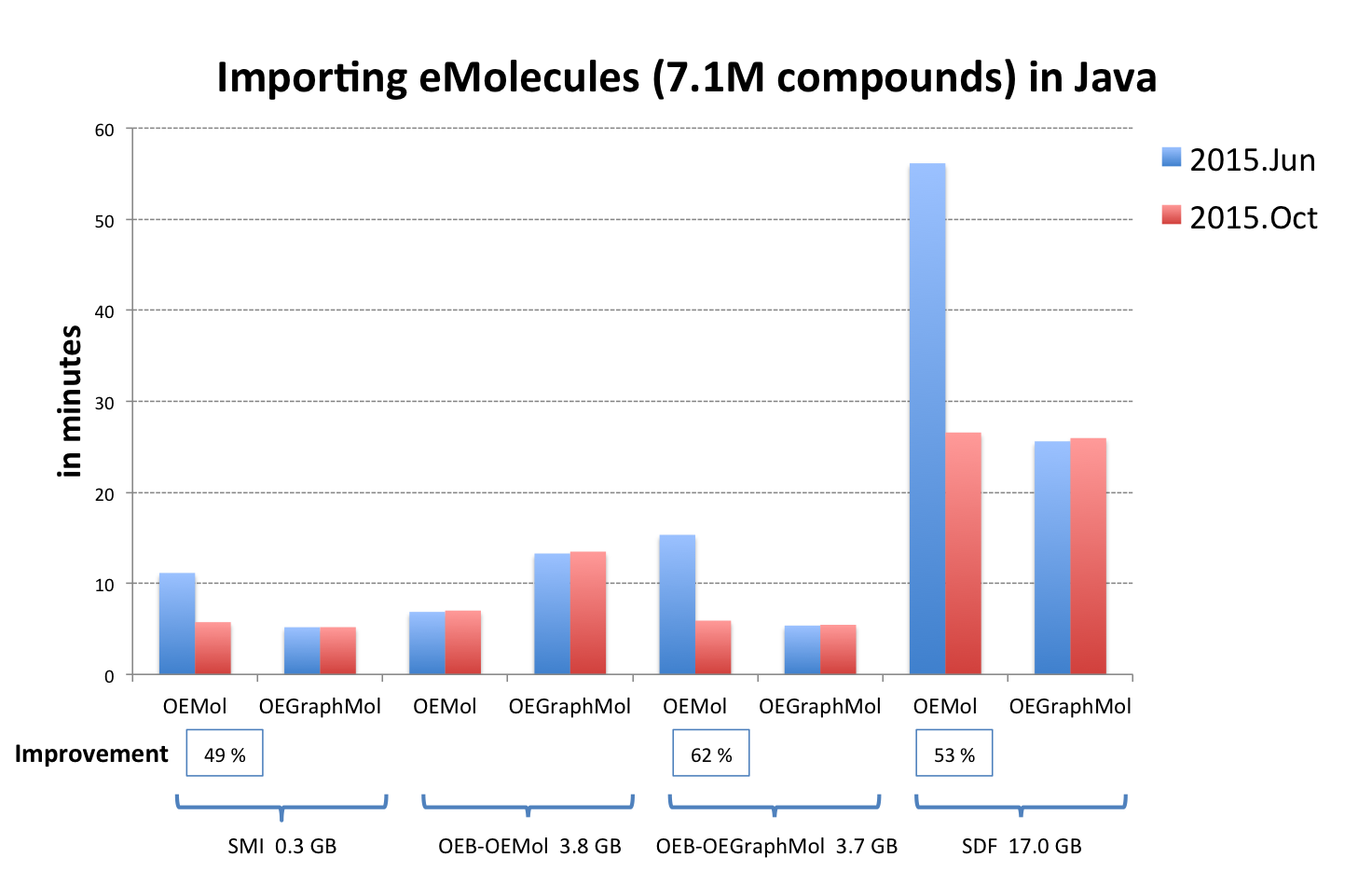
OEReadMoleculeinto an OEMol is now equivalent to reading into an OEGraphMol for the majority of use cases. To achieve this, it is now supported to callOEReadMoleculeon OEConfBase as returned byOEMCMolBase::GetActive. See table Performance improvement of importing molecules from various file formats below that shows the improvements by supported languages.
|
|
|
Note
OEB-OEMol - OpenEye OEBinary file format storing molecules as OEMol (multi-conformer molecule representation)
OEB-OEGraphMol - OpenEye OEBinary file format storing molecules as OEGraphMol (single-conformer molecule representation)
The performance of reading V2000 MDL MOL files has been significantly improved. See table Performance improvement of importing molecules from various file formats above.
The performance of 2D coordinate generation has been improved by as much as 33% in some cases.
OEIsMember::AddandOEIsMemberPtr::Addmethods have been added.Added new
OESMILESFlag::AllBondsoutput Flavor for SMILES generation. When set all bonds will be explicit in the generated output SMILES.A new
OEIsSDDataFormatfunction that can return whether the given file format supports SD data information has been added.OEGetMDLDimensionFromCoordsand OESetMDLDimensionFromCoords function have been added.OEMMFFTypeResult class was added along with a new
OEMMFFAtomTypeoverloaded function that provides better support for the wrapped languages (Python, Java and C#).213 new ring templates have been added to the built-in ring dictionary.
Example of the New 2D Ring Templates Carbon Skeleton
Specified Cis/Trans
The performance of reading
OEFormat::CDXfiles has been improved.SetTitlemethod of OEConfBase class is now slightly faster.
Major bug fixes
The memory subsystem in the OpenEye Toolkits has been revamped to allow more types of objects to pass safely between operating system threads by default. The following objects are no longer affected by
OESetMemPoolMode:The upgrade fixes crashes observed in the OEMolProp TK and OEDocking TK in multi-threaded web servers. A more thorough description of what can and can’t be done with threads with the OpenEye Toolkits can be found in the recently updated Multi-Threading chapter.
Care was taken to ensure the same or better level of performance with the new system. However, if you experience a performance degradation when switching to the Toolkit 2015.Oct release, please do not hesitate to contact support@eyesopen.com.
Previously,
OEMolBase::NewAtomwould not copy coordinates when the molecule was theOEMolBaseType::OEMiniMolimplementation. This problem has been fixed.OEMCMolBase::GetMCMolTitlewill now be consistent when reading single-conformer file formats. Previously, reading SMILES and CDX files would set the title on the parent OEMCMolBase, but MOL2, CSV, SDF, and single-conformer OEB file formats would not. All these file formats will now initialize the parent OEMCMolBase title allowing more consistent behavior when annotating conformer titles (warting) across all file formats. The conformers can be warted, leaving the parent title intact as well.OEChem::OEConfBase::operator boolwill now conform better to the OEMolBase API by returning whether the molecule contains atoms. The previous behavior was odd and unused, returning the state of the coordinates in the conformer.OEChem::OEConfBase::Clearwill now be consistent with theOEMolBase::ClearAPI effectively clearing away all generic data, SD data, atoms, and bonds from the parent OEMCMolBase.OEPerceiveResidueswill no longer crash when a thread has a small stack size and the molecule is very large.OEAddMolswill no longer crash when used onOEMolBaseType::OEMiniMolmolecule implementations.OEBondBase::Clearwill now clear generic data forOEMolBaseType::OEMiniMolmolecule implementations.The following changes were made to improve the support for
OEFormat::CDXfile format:The problem that caused 2D coordinates to be flipped resulting in flipped atom stereo after round-tripping has been fixed.
Hydrogen counts for charged aromatic atoms such as
[cH-]1cccc1have been fixed.OEBondStereo::WavyandOEBondStereo::DoubleEitherbonds are now correctly handled.
Minor bug fixes
The following methods of the OEAtomBase class are deprecated and will throw a warning when called:
OEAtomBase::HasAlphaBetaUnsat(use OEHasAlphaBetaUnsat predicate instead)
Previously, when the function
OEParseSmileswas called with a non-empty molecule, the hydrogen counts of the previously-existing atoms would be erroneously modified to satisfy normal valences. Now, the existing atom hydrogen counts are untouched.The function
OEReadMoleculewill now attempt to read malformed Rgroup atom information present in the atom block. Although an atom symbol ofR1in the atom block may seem reasonable, it does not strictly conform to the CTfile specification. TheOEWriteMoleculefunction will never export information in this form, but an attempt will be made to read malformed information on input.The function
OEReadMoleculewill no longer emit a warning when encountering the atom symbol*with CTfile format input, Instead, it will be treated as aOEElemNo::Duatom. This is in keeping with the return value fromOEGetAtomicNumfor*and a desire to reduce the number of warnings fromOEReadMolecule.The function
OEReadMoleculewill now read Rgroup atom label information from V3000 format input. Previously this information was ignored.The function
OEReadMoleculewill now make a more concerted effort to read malformed DOS line endings from input (text) files. This fix was added to oeistream.Trailing white spaces are now removed from SMILES titles.
When calling the
OE3DToInternalStereofunction, the bond stereo is perceived from 3D even if the atom stereo perception fails.Coordinates in MDL MOL files that cannot be parsed as a real floating point number will now be
0.0as per the MDL specification. Previously, the string “NaN” would actually translate to the floating pointNaNbinary representation.oemolistream::GetConfTestis nowconstcorrect. Theconst-nessof the underlying OEConfTestBase is no longer disregarded.oemolistream::GetFileNamewill no longer return an empty string when oemolistream constructor was called on a real file.During substructure search, the warning:
OESubSearch::SingleMatch() is unable to match bond stereo in the target for patterns with bond stereo, call OEPrepareSearch on the target firstwas sometimes incorrectly thrown. The underlying stereo perception has been revamped.OESubSearch no longer throws the following message in case when
OEPrepareSearchis invoked on the target molecule.OESubSearch::SingleMatch() is unable to match hybridization in the target for patterns with set hybridization, call OEPrepareSearch on the target first.The OEUniMolecularRxn has been improved to reduce the number of odd valence and broken aromatic ring results from transformations.
OEPerceiveChiralnow sets the relevant perception flag for empty molecules.
OEReadMolFromStringandOEWriteMolToStringfunctions now accept file format strings without the.prefix. So both “sdf” and “”.sdf”” will be interpreted as theOEFormat::SDFfile format.
OEIFlavor::SMI::StrictandOEIFlavor::SMI::Canonflavor combination is now valid for theOEFormat::SMIfile format.OE3DToAtomStereoandOE3DToBondStereofunctions remove the stereo property of an atom (or bond), respectively, if it is not chiral or if its stereo configuration can not be determined from the 3D.OEAtomBase::SetStereoandOEBondBase::SetStereomethods now remove the stereo generic data from the atoms (or bonds) when calling with theOEAtomStereo::UndefinedorOEBondStereo::Undefinedvalue, respectively.OEHasAtomStereoHydrogenswas fixed, it returnstruefor a tetrahedral atom with specified stereo and one explicit hydrogen neighbor.OEHasBondStereoHydrogenswas fixed, it returnstruefor an atom that belongs to a cis/trans double bond with one explicit hydrogen neighbor.OEReadOEBFilecan now be successfully used to read and round-trip an empty molecule (no atoms) to an OEB file.OEReadMoleculewill still automatically skip molecules with no atoms.OEBReadDataLengthnow takes a default parameter for the maximum length.OEFormat::Defaultnow has the same numerical value asOEFormat::SMIinstead ofOEFormat::ISM. As of OEChem TK 2.0.0 (the 2014.Feb release),OEFormat::SMIandOEFormat::ISMare functionally equivalent in OEChem TK’s file handling, so this is mostly a cosmetic change for consistency in that effort.Explicit hydrogens created by parsing stereo in SMILES strings is now capped to a maximum of eight explicit hydrogens. Previously, a SMILES such as
[C@@H1000000000]would seem to make OEChem TK hang indefinitely as it tried to create all those explicit hydrogens.OEMaskGridByCoordsAndRadiihas been corrected to fix a bug introduced in OEGrid 1.5.0 that produced grids of the wrong size.The function
OEReadMoleculewill now simply ignore the V3000 highlight collection information that was previously causing a read error.Previously, the function
OEWriteMoleculecould emit lines exceeding 80 chars in V3000 format for structures containing many stereocenters. This is in violation of the CTfile specification and has been corrected.An obscure issue causing a crash in OESubSearch or
OEQMolBase::BuildExpressionsfor imines and related queries with cis/trans parity and explicit hydrogens has been corrected, e.g.,[H]/N=C/C.
Documentation changes
rmsd example has been added to generate RMSD alignment for multi-conformer molecules.
Links to the CTfile format document from Biovia (previously Accelrys, Inc.) has been updated. Note that registration is required to download this document.
OE2DRingDictionary::AddRingsreturn value is now documented.
OEBio 2.0.5
New features
OEBio::OEFragmentNetwork class is now exposed in order to allow the investigation of the protein-ligand interactions perceived by OEDocking TK and visualized by Grapheme TK. The current API is read only - i.e., the user cannot build a fragment network.
The fragment network (OEBio::OEFragmentNetwork) is a container of typed molecules. The following classes are available to classify molecules in a fragment network:
OEBio::OEComponentTypeBase abstract class
OEBio::OEProteinComponentType concrete type that identifies protein molecule of complexes.
OEBio::OELigandComponentType concrete type that identifies ligand molecule of complexes.
A fragment (OEBio::OEFragment) is a set of atoms of a molecule that is stored in the fragment network.
A fragment connection (OEBio::OEFragmentConnection) is a typed link between two fragments of the network. The following classes have been added to handle interactions perceived by OEDocking TK (see also
OEAddDockingInteractionsfunction):OEBio::OEFragmentConnectionTypeBase abstract class
OEBio::OEFragmentContactInteraction
OEBio::OEFragmentHBondInteraction along with the related OEBio::OEFragmentHBondType namespace
OEBio::OEFragmentChelatorInteraction along with the related OEBio::OEFragmentChelatorType namespace
The following fragment connection predicates have been added:
OEBio::OEHasConnectionType
OEBio::OEHasConnection
OEBio::OEHasResidueConnection
OEBio::OEIsChelatorInteraction
OEBio::OEIsHBondInteraction
OEBio::OEIsInterConnection
OEBio::OEIsIntraConnection
The following functions have been added:
OEIsValidActiveSitefunctionOEBio::OEGetConnections function
The database of non-ligand residue types maintained by the OEResidueCategoryData object contained within an OESplitMolComplexOptions has been curated. Only about 40 percent of the entries previously listed under the category
OEMolComplexFilterCategory::Miscremain, the remaining entries were recognized to be obscure ligands or were moved to other categories, such asOEMolComplexFilterCategory::CofactorAndLigand. The categoryOEMolComplexFilterCategory::DNA_RNAhas been expanded to include the more obscure RNA bases.Setting modelNum to 0 in the
OESplitMolComplexOptions::ResetFiltersmethod of OESplitMolComplexOptions now selects all models.Protein Data Bank biological assembly files, with extensions like
.pdb1.gz, can now be read. These files use model numbers to refer to each part of an assembly (model numbers are also used to distinguish NMR models). By default, each model will be loaded as a separate molecule. See splitmolcomplex for an example of how to remove theENDMflavor in order to load all models from a Protein Data Bank file into a single molecule.
Minor bug fixes
A problem that may have affected which residues were selected when the option method
OESplitMolComplexOptions::SetSeparateResidueswas used to select binding site residues has been fixed.
Documentation changes
printinteractions example has been added to list protein-ligand interactions perceived by OEDocking TK.
The Stereochemistry Perception chapter has been rewritten for the sake of clarity.
Examples splitmolcomplex and splitmolcomplexfrags have been updated to support the reading of multiple models from a
PDBfile.
OESystem 2.0.5
New features
OERandom::GetSeedhas been added to return the current state of the OERandom class enabling the restart of the random number generator with the given seed.OEGridSizeMultiplyhas been added to safely multiply grid dimensions together while ensuring that it will not cause the value to overflow the integer type.OEOwnedPtr is now move-constructable for any C++11 enabled compiler. OpenEye’s first foray into supporting C++11 directly in our APIs. C++11 will eventually provide broader usage of smart pointers across the OpenEye Toolkits API.
Minor bug fixes
OEAnnotation can now be successfully round-tripped through OEB.
Python-specific changes
HasUnsignedIntandGetUnsignedIntmethods have been added to the OEInterface class Python wrapping to allow accessing command line parameters with!TYPE unsigned.
C#-specific changes
HasUnsignedIntandGetUnsignedIntmethods have been added to the OEInterface class C# wrapping to allow accessing command line parameters with!TYPE unsigned.
OEPlatform 2.0.5
New features
OEGetAbsolutePathis a new function that returns the full absolute path of a file.OEThread::SetStackSizehas been added to allow controlling the maximum amount of stack size that the operating system thread will have when it is started withOEThread::Start.If a toolkit program fails due to a license error, the paths of any license file(s) actually read will be output for diagnostic purposes.
Major bug fixes
OEPlatform::OEMallocawill no longer cause crashes when attempting to allocate amounts of memory near the maximum allowablesize_t.std::bad_allocwill now be thrown instead in these situations. SinceOEPlatform::OEMallocais actually a C-preprocessor macro, this change may result in user code throwing additional compiler warnings when signed integer types like ‘int’ are passed toOEPlatform::OEMalloca. We recommend that users change this code to usesize_tto avoid these warnings.
Minor bug fixes
oepstreamclass has been removed since it never worked as originally intended.OEUncompressandOECompresscan now takesize_tinstead ofunsigned intallowing use on data in excess of 4GB on 64-bit machines.
Documentation changes
OEOnce example code in the documentation will now actually compile. Previously, the code would actually crash prior to the OESubSearch memory pool refactoring of this release, if any other thread besides the
mainthread called the function first.
C++-specific changes
OpenEye Toolkits header files should no longer contain the C++11 deprecated “register” keyword causing compilations with
-Werrorto fail.oestream and derived classes will no longer allow implicit conversion to
intandunsigned int. This avoids surprising behavior like the following from compiling:oeisstream is1; oeisstream is2; bool what = (is1 == is2);
OEGrid
New features
OEGrid TK now features more efficient use of memory while reading grids from files. Previously, several unnecessary memory allocations had sometimes been performed during the read process.
OEGrid TK grids are now officially capped to 4GB of memory since the API is designed around
unsigned int.
Major bug fixes
OEScalarGrid and all other grid types will no longer crash when the dimensions would cause overflow of an
unsigned int. The grid types will now throwstd::bad_allocif attempting to make the grid memory larger than 4GB.