Annotating Atoms and Bonds
The Grapheme TK provides a framework to annotate atoms and bonds i.e.
mark them based on their properties.
The following Listing 1 example shows
how to mark \(sp^2\) and \(sp^3\) hybridized atoms of
depicted molecules.
After preparing the molecule for 2D depiction and calculating the
atom hybridizations, the molecule display object is created
(OE2DMolDisplay) that stores the depiction
information of a molecule
The OEAddGlyph function is then called to
add glyph to those atoms for which the given
OEIsAtomHybridization functor returns true.
In this example, the built-in OEAtomGlyphCircle
class is used to mark atoms by drawings a circle around them with a
specific style (OECircleStyle).
Finally, the image is written out to a file by calling the
OEWriteImage function.
The image created by Listing 1 example
is shown in
Figure: Example of atom annotation.
Listing 1: Example of using a built-in atom annotation style
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "c1cc(N)cc(S(=O)(=O)O)c1")
oechem.OEAssignHybridization(mol)
oedepict.OEPrepareDepiction(mol)
opts = oedepict.OE2DMolDisplayOptions(350, 250, oedepict.OEScale_AutoScale)
opts.SetTitleLocation(oedepict.OETitleLocation_Hidden)
disp = oedepict.OE2DMolDisplay(mol, opts)
sp2pen = oedepict.OEPen(oechem.OEWhite, oechem.OEBlueTint, oedepict.OEFill_Off, 1.5)
glyphSP2 = oegrapheme.OEAtomGlyphCircle(sp2pen, oegrapheme.OECircleStyle_Sun, 1.2)
oegrapheme.OEAddGlyph(disp, glyphSP2, oechem.OEIsAtomHybridization(oechem.OEHybridization_sp2))
sp3pen = oedepict.OEPen(oechem.OEWhite, oechem.OEPinkTint, oedepict.OEFill_Off, 1.5)
glyphSP3 = oegrapheme.OEAtomGlyphCircle(sp3pen, oegrapheme.OECircleStyle_Eyelash, 1.2)
oegrapheme.OEAddGlyph(disp, glyphSP3, oechem.OEIsAtomHybridization(oechem.OEHybridization_sp3))
oedepict.OERenderMolecule("AnnotateAtomPredicate.png", disp)
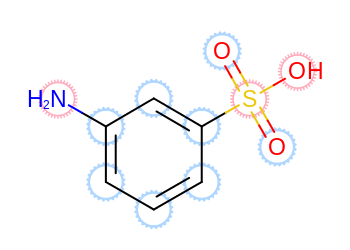
Example of atom annotation
See also
OEAddGlyphfunctionOEAtomGlyphCircle class
OECircleStylenamespaceOE2DMolDisplay class and
OEWriteImagefunction in the OEDepict TK manual
Similarly, the following example shows how to annotate bonds.
The bonds being highlighted are specified by the bond predicate passed
to the OEAddGlyph function.
For each bond the predicate returns true, i.e. if it is a
rotatable bond, the OEBondGlyphArrow.RenderGlyph
method is invoked that draws an arrow across the middle of the specific bond.
The image created by Listing 2 is shown in
Figure: Example of bond annotation.
Listing 2: Example of using a built-in bond annotation style
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "c1cc(NCC)cc(CS(=O)(=O)O)c1")
oedepict.OEPrepareDepiction(mol)
opts = oedepict.OE2DMolDisplayOptions(400, 250, oedepict.OEScale_AutoScale)
opts.SetTitleLocation(oedepict.OETitleLocation_Hidden)
disp = oedepict.OE2DMolDisplay(mol, opts)
pen = oedepict.OEPen(oechem.OEDarkPurple, oechem.OEDarkPurple, oedepict.OEFill_Off, 2.0)
glyph = oegrapheme.OEBondGlyphArrow(pen, 0.5)
oegrapheme.OEAddGlyph(disp, glyph, oechem.OEIsRotor())
oedepict.OERenderMolecule("AnnotateBondPredicate.png", disp)
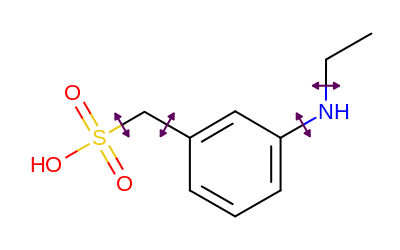
Example of bond annotation
The following table lists the customizable bond glyphs that are currently available in Grapheme TK.
|
|
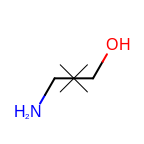
|
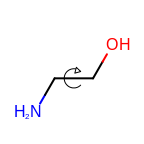
|
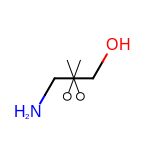
|
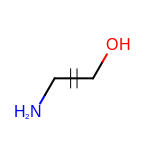
|
|
|
See also
OEAddGlyphfunction
The last example shows how to depict non-boolean properties by implementing
a user-defined annotation style.
User-defined atom and bond annotations can be implemented by deriving
from the OEAtomGlyphBase and the
OEBondGlyphBase abstract classes and implementing
the OEAtomGlyphBase.RenderGlyph and the
OEBondGlyphBase.RenderGlyph methods,
respectively.
In the Listing 3 example, after
calculating the MMFF charges, the atoms are annotated by their charge
using the OELinearColorGradient class that
interpolates colors. Atoms with negative and positive charge are
highlighted by red and blue colors, respectively.
The image created by Listing 3 is shown in
Figure: Example of user-defined annotation.
Listing 3: Example of user-defined annotation
class ColorCharge(oegrapheme.OEAtomGlyphBase):
def __init__(self, cg):
oegrapheme.OEAtomGlyphBase.__init__(self)
self.colorg = cg
def RenderGlyph(self, disp, atom):
adisp = disp.GetAtomDisplay(atom)
if adisp is None or not adisp.IsVisible():
return False
charge = atom.GetPartialCharge()
if charge == 0.0:
return True
color = self.colorg.GetColorAt(charge)
pen = oedepict.OEPen()
pen.SetForeColor(oechem.OEColor(color))
color.SetA(100)
pen.SetBackColor(oechem.OEColor(color))
pen.SetFill(oedepict.OEFill_On)
radius = disp.GetScale() / 2.5
layer = disp.GetLayer(oedepict.OELayerPosition_Below)
oegrapheme.OEDrawCircle(layer, oegrapheme.OECircleStyle_Simpson,
adisp.GetCoords(), radius, pen)
return True
def CreateCopy(self):
return ColorCharge(self.colorg).__disown__()
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "Cc1cc(cc(c1[N+](=O)[O-])F)[N+]#C")
oechem.OEMMFFAtomTypes(mol)
oechem.OEMMFF94PartialCharges(mol)
oedepict.OEPrepareDepiction(mol)
opts = oedepict.OE2DMolDisplayOptions(350, 250, oedepict.OEScale_AutoScale)
opts.SetAtomColorStyle(oedepict.OEAtomColorStyle_WhiteMonochrome)
opts.SetTitleLocation(oedepict.OETitleLocation_Hidden)
disp = oedepict.OE2DMolDisplay(mol, opts)
coloranion = oechem.OEColorStop(-1.0, oechem.OEColor(oechem.OEDarkRed))
colorcation = oechem.OEColorStop(+1.0, oechem.OEColor(oechem.OEDarkBlue))
colorg = oechem.OELinearColorGradient(coloranion, colorcation)
colorg.AddStop(oechem.OEColorStop(0.0, oechem.OEColor(oechem.OEWhite)))
colorcharge = ColorCharge(colorg)
oegrapheme.OEAddGlyph(disp, colorcharge, oechem.OEIsTrueAtom())
oedepict.OERenderMolecule("AnnotatePartialCharge.png", disp)
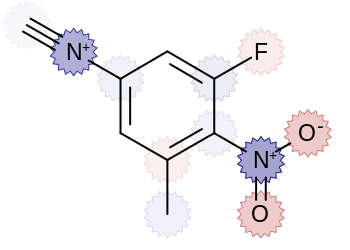
Example of user-defined annotation
See also
OEAtomGlyphBase class
OEBondGlyphBase class
OEAddGlyphfunctionOELinearColorGradient class