GraphemeTM TK 1.3.3
New features
A new function,
OERenderContactMap, has been added that generates interactive SVG images. Clicking on any residue circle will highlight those ligand atoms that interact with the given residue.
click on any residue
Example of interactive SVG image generated by OERenderContactMap (PDB: 4HWR)
Preliminary API
A new Preliminary API has been added to mark up the residue circles in SVG images generated by the
OERenderActiveSitefunction:OE2DActiveSiteDisplayOptions.SetResidueSVGMarkupFunctorandOE2DActiveSiteDisplayOptions.GetResidueSVGMarkupFunctormethodsOEResidueSVGMarkupBase base class and OEResidueSVGNoMarkup and OEResidueSVGStandardMarkup concrete classes
Major bug fixes
Several problems in the
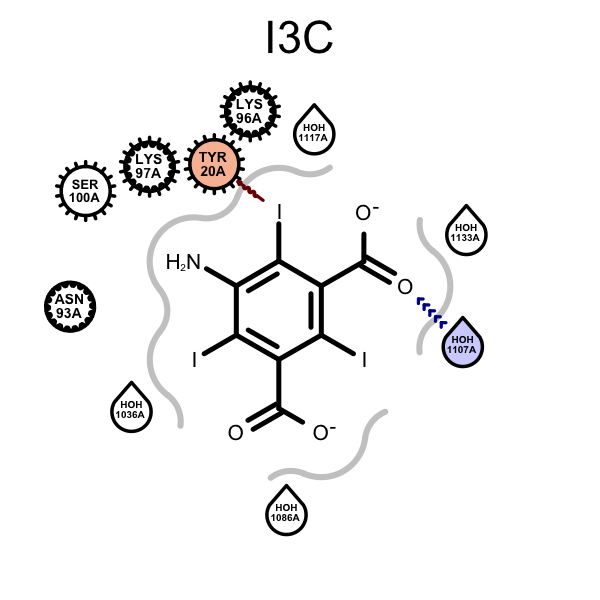
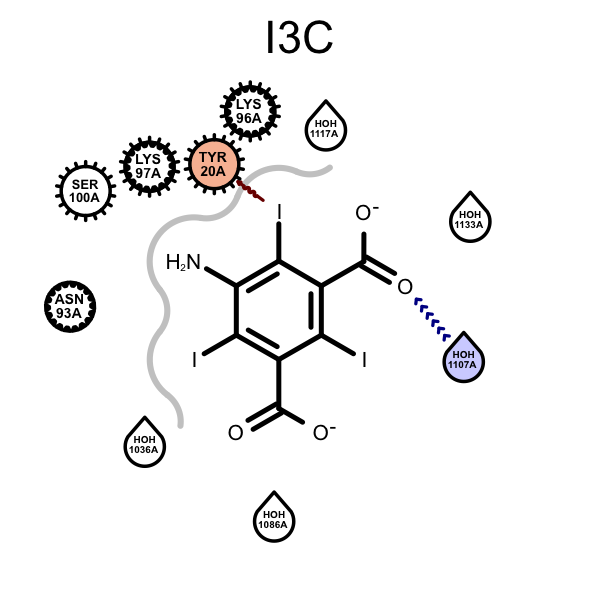
OERenderActiveSitefunction have been fixed:When depicting protein-ligand interactions, grey lines represent the shape of the binding pocket. The absence of lines indicates solvent-accessible regions. In order visualize the solvent-accessible areas more realistically, waters are now ignored when calculating the molecule surfaces that determine the 2D representation (compare images below).
The rare case where the ligand is completely surrounded by water is now handled correctly.
The following heuristics are now used to improve the positioning of residue glyphs around the ligand:
Waters are more likely to be positioned in solvent-exposed regions.
Other residues are only positioned in solvent-exposed regions if it results in a more visually pleasing layout.
Example of visualizing solvent-accessible regions and water positioning (PDB: 3E3D) 2017.Feb
2017.Jun
Minor bug fixes
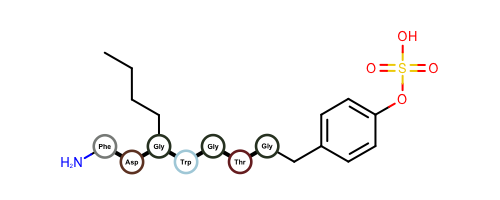
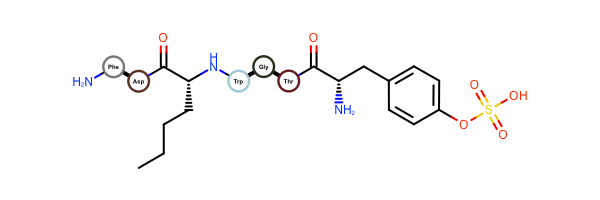
OEDrawPeptidehas been fixed and now allows only amide and disulfide bonds when identifying standard amino acids. No alpha-carbon substitutions are allowed.Example of visualizing peptides with OEDrawPeptide 2017.Feb
2017.Jun
Documentation changes
A new Python Cookbook Examples section has been added.