Searching fast fingerprint database
A program that initializes a fast fingerprint database with the fingerprint file generated from the previous example and then performs a fingerprint search to identify molecules which are similar to a given query molecule.
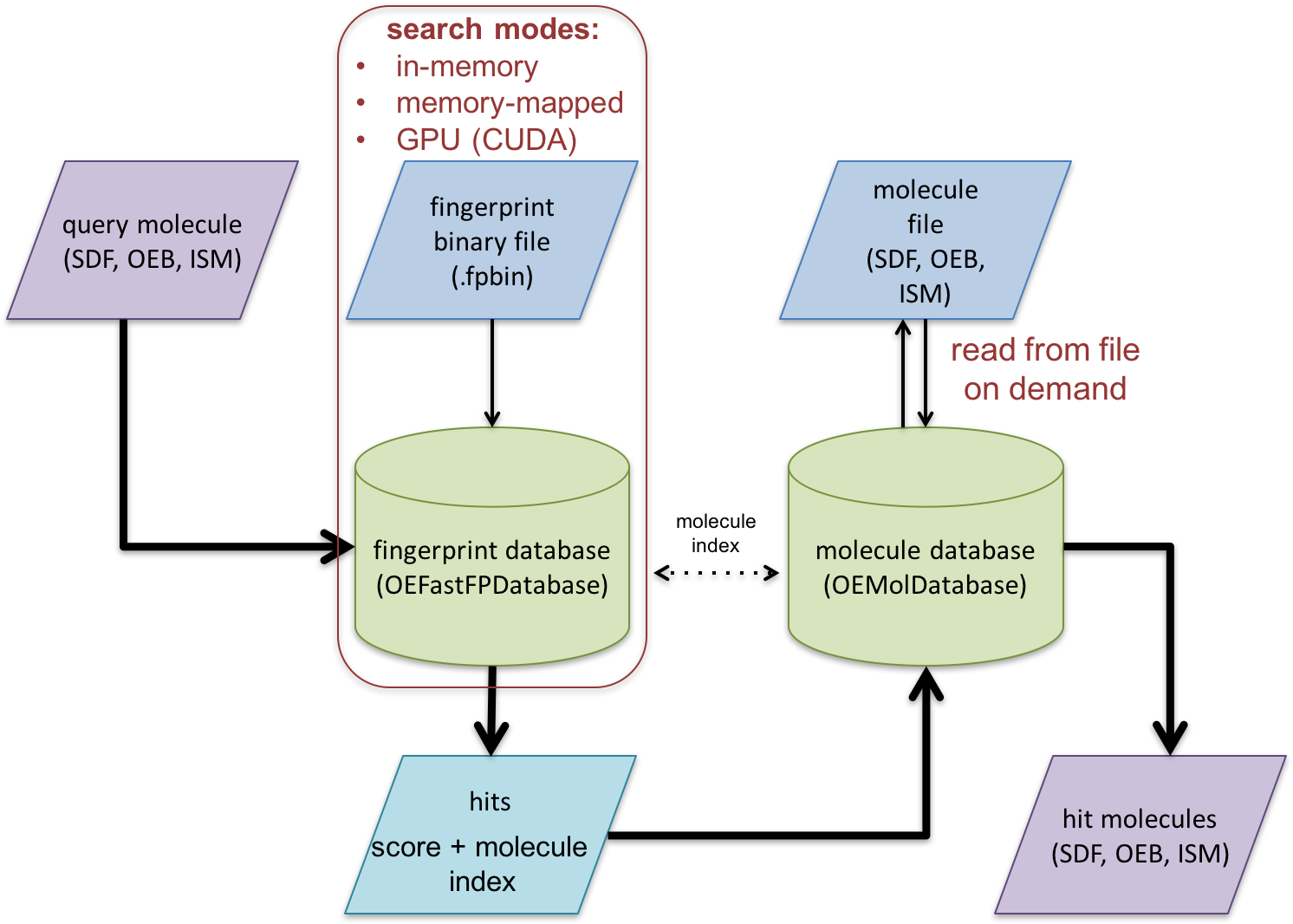
Schematic representation of fast fingerprint search process
See also
Example code in the Generating fingerprint file for fast fingerprint search section
Rapid Similarity Searching of Large Molecule File OpenEye Python Cookbook example
Command Line Interface
A description of the command line interface can be obtained by executing the program with the –help argument.
prompt> python searchfastfp.py --help
will generate the following output:
Simple parameter list
fingerprint database options
-memorytype : Fingerprint database memory type
fingerprint database search options
-cutoff : Similarity cutoff value
-descending : Order of similarity scores
-limit : Maximum number of similarity scores
-simfunc : Similarity measure
input/output options
-fpdbfname : Input fast fingerprint database filename
-molfname : Input molecule filename
-out : Output molecule filename
-query : Input query filename
Code
Download code
#!/usr/bin/env python
# (C) 2022 Cadence Design Systems, Inc. (Cadence)
# All rights reserved.
# TERMS FOR USE OF SAMPLE CODE The software below ("Sample Code") is
# provided to current licensees or subscribers of Cadence products or
# SaaS offerings (each a "Customer").
# Customer is hereby permitted to use, copy, and modify the Sample Code,
# subject to these terms. Cadence claims no rights to Customer's
# modifications. Modification of Sample Code is at Customer's sole and
# exclusive risk. Sample Code may require Customer to have a then
# current license or subscription to the applicable Cadence offering.
# THE SAMPLE CODE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND,
# EXPRESS OR IMPLIED. OPENEYE DISCLAIMS ALL WARRANTIES, INCLUDING, BUT
# NOT LIMITED TO, WARRANTIES OF MERCHANTABILITY, FITNESS FOR A
# PARTICULAR PURPOSE AND NONINFRINGEMENT. In no event shall Cadence be
# liable for any damages or liability in connection with the Sample Code
# or its use.
#############################################################################
# Searching fast fingerprint database
#############################################################################
import sys
from openeye import oechem
from openeye import oegraphsim
def main(argv=[__name__]):
itf = oechem.OEInterface()
oechem.OEConfigure(itf, InterfaceData)
defopts = oegraphsim.OEFPDatabaseOptions(10, oegraphsim.OESimMeasure_Tanimoto)
oegraphsim.OEConfigureFPDatabaseOptions(itf, defopts)
oegraphsim.OEConfigureFPDatabaseMemoryType(itf)
if not oechem.OEParseCommandLine(itf, argv):
return 0
qfname = itf.GetString("-query")
mfname = itf.GetString("-molfname")
ffname = itf.GetString("-fpdbfname")
ofname = itf.GetString("-out")
# initialize databases
timer = oechem.OEWallTimer()
timer.Start()
ifs = oechem.oemolistream()
if not ifs.open(qfname):
oechem.OEThrow.Fatal("Cannot open input file!")
query = oechem.OEGraphMol()
if not oechem.OEReadMolecule(ifs, query):
oechem.OEThrow.Fatal("Cannot read query molecule!")
moldb = oechem.OEMolDatabase()
if not moldb.Open(mfname):
oechem.OEThrow.Fatal("Cannot open molecule database!")
memtype = oegraphsim.OEGetFPDatabaseMemoryType(itf)
fpdb = oegraphsim.OEFastFPDatabase(ffname, memtype)
if not fpdb.IsValid():
oechem.OEThrow.Fatal("Cannot open fingerprint database!")
nrfps = fpdb.NumFingerPrints()
memtypestr = fpdb.GetMemoryTypeString()
ofs = oechem.oemolostream()
if not ofs.open(ofname):
oechem.OEThrow.Fatal("Cannot open output file!")
if not oegraphsim.OEAreCompatibleDatabases(moldb, fpdb):
oechem.OEThrow.Fatal("Databases are not compatible!")
oechem.OEThrow.Info("%5.2f sec to initialize databases" % timer.Elapsed())
fptype = fpdb.GetFPTypeBase()
oechem.OEThrow.Info("Using fingerprint type %s" % fptype.GetFPTypeString())
opts = oegraphsim.OEFPDatabaseOptions()
oegraphsim.OESetupFPDatabaseOptions(opts, itf)
# search fingerprint database
timer.Start()
scores = fpdb.GetSortedScores(query, opts)
oechem.OEThrow.Info("%5.2f sec to search %d fingerprints %s"
% (timer.Elapsed(), nrfps, memtypestr))
timer.Start()
nrhits = 0
hit = oechem.OEGraphMol()
for si in scores:
if moldb.GetMolecule(hit, si.GetIdx()):
nrhits += 1
oechem.OESetSDData(hit, "Similarity score", "%.2f" % si.GetScore())
oechem.OEWriteMolecule(ofs, hit)
oechem.OEThrow.Info("%5.2f sec to write %d hits" % (timer.Elapsed(), nrhits))
return 0
InterfaceData = """
!BRIEF [-query] <molfile> [-molfname] <molfile> [-fpdbfname] <fpfile> [-out] <molfile>
!CATEGORY "input/output options"
!PARAMETER -query
!ALIAS -q
!TYPE string
!REQUIRED true
!KEYLESS 1
!VISIBILITY simple
!BRIEF Input query filename
!END
!PARAMETER -molfname
!ALIAS -mol
!TYPE string
!REQUIRED true
!KEYLESS 2
!VISIBILITY simple
!BRIEF Input molecule filename
!END
!PARAMETER -fpdbfname
!ALIAS -fpdb
!TYPE string
!REQUIRED true
!KEYLESS 3
!VISIBILITY simple
!BRIEF Input fast fingerprint database filename
!END
!PARAMETER -out
!ALIAS -o
!TYPE string
!REQUIRED true
!KEYLESS 4
!VISIBILITY simple
!BRIEF Output molecule filename
!END
!END
"""
if __name__ == "__main__":
sys.exit(main(sys.argv))
See also
OEFPDatabaseOptions class
OEConfigureFPDatabaseOptionsandOESetupFPDatabaseOptionsfunctionsOEConfigureFPDatabaseMemoryTypeandOEGetFPDatabaseMemoryTypefunctionOEMolDatabase class in OEChem TK manual
OEFastFPDatabaseMemoryTypenamespaceOEFastFPDatabase class
OEAreCompatibleDatabasesfunction