Version 2.0.4
OEChem 2.0.4
New features
The following API have been added to generate and use 2D ring dictionaries:
OE2DRingDictionary class
OEIsValid2DRingDictionaryfunctionOEWrite2DRingDictionaryfunctionOEInit2DRingDictionaryfunction
The following sections provide examples to generate and utilize user-defined 2D ring dictionaries:
See also
ringdict2pdf example in OEDepict TK will generate a multi-page PDF report of a 2D ring dictionary.
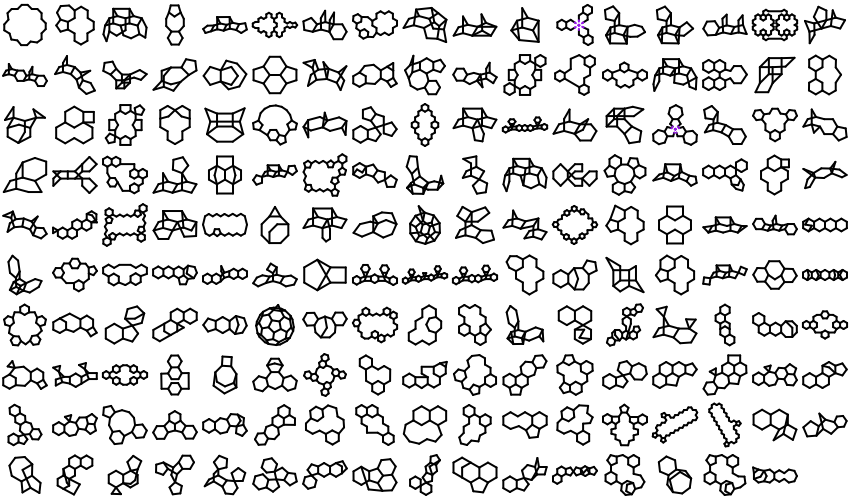
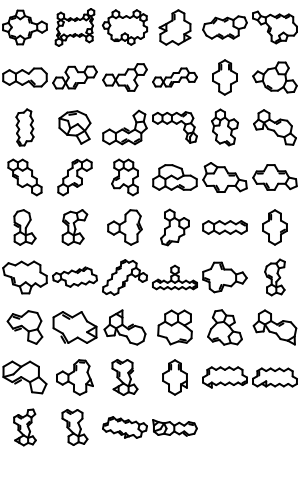
221 new ring templates have been added to the built-in ring dictionary.
Example of the New 2D Ring Templates Pure Carbon Skeleton Ring Templates
Ring Templates with Specified CIS/Trans Bonds
OEIFlavor.MOL2.Forcefieldflavor has been added to the MOL2 reader. This allows atom names in the MOL2 file to be interpreted as chemical elements while not using the second character if capitalized; for example, CA and CD are both carbon instead of calcium and cadmium. This will help readingOEFormat.MOL2files that contain forcefield information like partial charges.OEAddCustomFASTAResiduenow provides the ability to customizeOEFormat.FASTAinput files with unnatural nucleic acids.OEIFlavor.FASTA.CustomResiduesflavor is used to activate the new feature.OEIFlavor.FASTA.EmbeddedSMILESflavor has also been added to experiment with adding cyclizations to peptides using custom tethers. These features are currently considered experimental; please send feedback to support@eyesopen.com.
OEOFlavor.SDF.UnsetBad2DStereoandOEOFlavor.CDX.UnsetBad2DStereo, new output flavors for SDF and CDX output formats, have been added. These constants control behavior when generating 2D coordinates during output. When used, the writer will set the stereo to “undefined” for any misleading stereochemistry that may result from layout limitations.OESubsetMolhas a new overload that supports the use of OEAtomBondSet input. This can offer significant performance improvements over the predicate overloads since a full molecule iteration can be avoided.OEMolBase.ClearCoordshas been added to fully clear molecule coordinates, clear the dimension code, and reset perception flags.OEMolDatabase.WriteMoleculehas been added to automatically extract a molecule from the database and write it to the oemolostream. This function is optimized for performance to simply move bytes around if the input OEMolDatabase format and the output format match exactly.OEMolDatabase.GetMoleculeStringhas been added to allow direct access to the molecular record data.OEPRECompressandOESweepRotorCompressHydrogenshave been added and documented to allow advanced users to start experimenting with perfect-rotor-encoding OEB files to achieve higher rates of conformer compression. This feature is currently only available to the C++ toolkits.OEMDLGetValencehas been added to provide direct access to the MDL valence model. See the MDL Valence Model section for more information.The following free functions have been added for handling dummy atoms in forcefield calculations in special circumstances:
Currently, dummy atoms are only allowed in Szybki TK for non-bonded interactions.
OEAssignRadiifree function has been added to allow for a central entry point for any OpenEye Toolkit atomic radii assignment. This free function uses theOERadiiTypenamespace for choosing the type of radii to assign.
Major bug fixes
OEReadMoleculewill now correctly handle right-justified atomic symbols in MDL molfiles. These files are being generated by the RCSB PDB custom SDF file writer.OEPerceiveCIPStereo, the Cahn-Ingold-Prelog stereochemistry perception algorithm, has been rewritten to solve the following issues:Performance issues for large molecules have been resolved.
Incorrect chirality perception for certain isotopic molecules, detailed in the blog post R or S? Let’s Vote, has been corrected.
Incorrect chirality perception for stereocenters with both a hydrogen and either a deuterium or tritium attached has been fixed.
Incorrect chirality perception for stereocenters with both a hydrogen and a *atom (atomic number 0) has been fixed.
oemolistream.Setgzwith afalseargument called after a stream has finished reading molecules will no longer causeOEReadMoleculeto crash on subsequent calls. It will now returnfalseto indicate that the stream has finished.OEMolDatabase.Openwill now fail and throw a warning message if there is no disk space left for uncompressing compressed input files. In earlier releases, the operation would appear to succeed and lead to corrupted molecules past a certain point. Furthermore, the directory that OEMolDatabase uses for temporary files can now be controlled through theTEMPorTMPenvironment variables on Windows or theTMPDIRenvironment variable on Linux and OSX.OEMol will now work with the
V3000file format much more reliably. This was accomplished by making sure that multi-conformer molecules can handle groups appropriately. The OEGroupBase class is a container of atom and bond pointers inside an OEMCMolBase. The OEGroupBase class is currently used to store MDL-enhanced stereo information. Each conformer of the multi-conformer molecule has its own separate container. Now methods such asOEMolBase.GetGroupswill provide access to the groups of the active conformation.
Minor bug fixes
OEFormat.OEBformat reading will no longer cause a rare crash when dealing with large amounts of SD data, PDB data, or bond integer types.OEMatchBase now checks parameters to the overloaded versions of the
OESubsetMolfunction to ensure that the given match corresponds to source and destination molecules.OEMolBase.NewGroupmethod signatures now takeconst std::vectorof atom and/or bond pointers. Previously, these arguments were notconsteven though the vectors were not altered.OEReadMoleculewill now perform chirality perception for 0D MDL molfiles to ensure thatOEAtomBase.IsChiralandOEBondBase.IsChiralare correct.OEGetAtomicNumis now tolerant of leading spaces for the string argument passed in.
Java-specific changes
OEBinaryIOHandlerBase had accidentally been removed from Java in the 2014.Oct release. This broke the older idiom for preserving rotor-offset compression in OEB files using
OEInitDefaultHandler. The preferred API is to now useOEPreserveRotCompressdirectly on the molecule streams, avoiding the need to access the OEBinaryIOHandlerBase internals entirely.The
OEUnaryRoleSetPredicateFuncfunction was renamed toOEUnaryRoleSetBoolFuncfor the sake of consistency.OERoleSet predicates can now be created and customized in the same way as atom and bond predicates. OERoleSet predicates are an important part of customizing the new OEBio TK Splitting Macromolecular Complexes feature.
OERoles class has been added as a concrete implementation of the OERoleSet abstract base class.
Documentation changes
2D Coordinate Generation chapter has been added.
OEBio 2.0.4
New features
An API has been added for classifying and separating the various components of a macromolecular complex by their functional roles. See the Splitting Macromolecular Complexes section for more information.
OESplitMolComplexfunctionOEGetMolComplexComponentsfunctionOECountMolComplexSitesfunctionOESplitMolComplexOptions class
OEMolComplexCategorizer class
OEResidueCategoryData class
OEMolComplexFilter class
OEGetResiduesfunction has been added to allow iteration over all unique OEResidue objects in an OEMolBase.
Documentation changes
Protein Preparation chapter has been added to document the APIs for splitting macromolecular complexes into their components.
OESystem 2.0.4
New features
OEThrow.SetLevelandOEThrow.GetLevelrace conditions should be largely alleviated now.OEErrorHandler.SetLevelandOEErrorHandler.GetLevelwill now affect the error message level for the current thread only; all new threads launch with the default error level ofOEErrorLevel.Default(Infoor above).Warning
Multi-threaded OpenEye Toolkit programs may need to be updated to allow the desired output to be sent to standard error. However, the previous behavior was race-condition prone and difficult to get correct.
Minor bug fixes
OERandom constructor will no longer take an
unsigned intas input. Previously, theunsigned intwould be implicitly cast tobooland would control whether to time seed the constructor. However, most users actually intend to use anunsigned intas a random number seed meant for theOERandom.Seedmethod. This change should catch this common mistake at compile time.
Documentation changes
OEMath Functions chapter now contains more detailed descriptions of the functions.
Documentation has been added for the following classes:
OEPlatform 2.0.4
New features
OEFileTempPathwill now use the directory specified by theTMPDIRenvironment variable on Linux and OSX. This allows the user to control where OpenEye Toolkits will create temporary files. This is especially useful since OEMolDatabase can use a lot of temporary storage when dealing with compressed data files.
OEGrid 1.5.1
Minor internal changes have been made.