Aligning Molecules Based on Substructure Search
A program that aligns the molecules based on a substructure search initialized with the given SMARTS pattern.
Command Line Interface
A description of the command line interface can be obtained by executing the program with the –help argument.
prompt> python smartsalign2D.py --help
Will generate the following output:
Simple parameter list
input/output options :
-in : Filename of input molecules
-out : Output filename
-smarts : SMARTS for alignment match
Code
Download code
#!/usr/bin/env python
# (C) 2022 Cadence Design Systems, Inc. (Cadence)
# All rights reserved.
# TERMS FOR USE OF SAMPLE CODE The software below ("Sample Code") is
# provided to current licensees or subscribers of Cadence products or
# SaaS offerings (each a "Customer").
# Customer is hereby permitted to use, copy, and modify the Sample Code,
# subject to these terms. Cadence claims no rights to Customer's
# modifications. Modification of Sample Code is at Customer's sole and
# exclusive risk. Sample Code may require Customer to have a then
# current license or subscription to the applicable Cadence offering.
# THE SAMPLE CODE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND,
# EXPRESS OR IMPLIED. OPENEYE DISCLAIMS ALL WARRANTIES, INCLUDING, BUT
# NOT LIMITED TO, WARRANTIES OF MERCHANTABILITY, FITNESS FOR A
# PARTICULAR PURPOSE AND NONINFRINGEMENT. In no event shall Cadence be
# liable for any damages or liability in connection with the Sample Code
# or its use.
#############################################################################
# Aligns molecules to a smarts pattern
#############################################################################
import sys
from openeye import oechem
from openeye import oedepict
def main(argv=[__name__]):
itf = oechem.OEInterface(InterfaceData)
if not oechem.OEParseCommandLine(itf, argv):
oechem.OEThrow.Fatal("Unable to interpret command line!")
iname = itf.GetString("-in")
oname = itf.GetString("-out")
smarts = itf.GetString("-smarts")
qmol = oechem.OEQMol()
if not oechem.OEParseSmarts(qmol, smarts):
oechem.OEThrow.Fatal("Invalid SMARTS: %s" % smarts)
oechem.OEGenerate2DCoordinates(qmol)
ss = oechem.OESubSearch(qmol)
if not ss.IsValid():
oechem.OEThrow.Fatal("Unable to initialize substructure search!")
ifs = oechem.oemolistream()
if not ifs.open(iname):
oechem.OEThrow.Fatal("Cannot open input molecule file!")
ofs = oechem.oemolostream()
if not ofs.open(oname):
oechem.OEThrow.Fatal("Cannot open output file!")
if not oechem.OEIs2DFormat(ofs.GetFormat()):
oechem.OEThrow.Fatal("Invalid output format for 2D coordinates")
for mol in ifs.GetOEGraphMols():
oechem.OEPrepareSearch(mol, ss)
alignres = oedepict.OEPrepareAlignedDepiction(mol, ss)
if not alignres.IsValid():
oechem.OEThrow.Warning("Substructure is not found in input molecule!")
oedepict.OEPrepareDepiction(mol)
oechem.OEWriteMolecule(ofs, mol)
return 0
#############################################################################
# INTERFACE
#############################################################################
InterfaceData = '''
!BRIEF [-in] <input> [-smarts] <smarts> [-out] <output>
!CATEGORY "input/output options :"
!PARAMETER -in
!ALIAS -i
!TYPE string
!REQUIRED true
!KEYLESS 1
!VISIBILITY simple
!BRIEF Filename of input molecules
!END
!PARAMETER -smarts
!ALIAS -s
!TYPE string
!REQUIRED true
!KEYLESS 2
!VISIBILITY simple
!BRIEF SMARTS for alignment match
!END
!PARAMETER -out
!ALIAS -o
!TYPE string
!REQUIRED true
!KEYLESS 3
!VISIBILITY simple
!BRIEF Output filename
!END
!END
'''
if __name__ == "__main__":
sys.exit(main(sys.argv))
See also
Molecule Alignment chapter
OESubSearch class
OEGenerate2DCoordinatesfunctionOEIs2DFormatfunctionOEPrepareSearchfunctionOEPrepareAlignedDepictionfunctionOEPrepareDepictionfunction
Examples
prompt> python smartsalign2D.py -in test.sdf -out aligned.sdf -smarts "c1cc[c,n]c2c1ncc2" -out out.mol
Taking the output coordinates and generating the images with the mol2img program will produce the images shown in Table: Example of using the program to align molecule
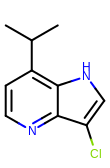
|
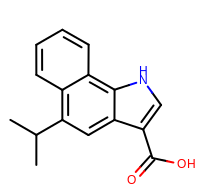
|
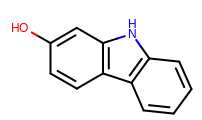
|
See also
Molecule Alignment chapter