OEPrepareAlignedDepiction
Link |
Description |
|---|---|
OEPrepareAlignedDepiction(fitmol, refmol, match, clearcoords, suppressH) |
aligns a molecule to the reference |
aligns a molecule to the reference according to options |
|
aligns a molecule to the reference based on maximum common substructure search |
|
aligns a molecule to the reference based on substructure search |
|
aligns a molecule to the reference based on matches |
bool OEPrepareAlignedDepiction(OEChem::OEMolBase &fitmol,
const OEChem::OEMolBase &refmol,
const OEChem::OEMatchBase &match,
bool clearCoords=true, bool suppressH=true)
Generates the 2D coordinates of a molecule by maximizing its overlap with a reference 2D molecule based on the given match between the two molecules.
- fitmol
The molecule being fitted to be aligned to the reference molecule.
- refmol
The reference molecule of which 2D coordinates are remain unchanged.
- match
The match determines the atoms of the fitmol that will be aligned to the atoms of the refmol. The match can be generated by hand, or with any of OEChem TK‘s matching algorithms such as OESubSearch or OEMCSSearch.
- clearcoords
If true, than the 2D coordinates of the fitted molecule are re-calculated by calling the
OEPrepareDepictionfunction. After generating the coordinates, the alignment between the two molecules is maximized by rotating the single bonds of the fitted molecule while avoiding atom clashes. See example (C) in Figure: Example of the molecule alignment.If clearcoords is false and the fitmol has 2D coordinates than these coordinates are kept. In this case only 2D rotation and translation of the fitted molecule is allowed (i.e. no rotations around bonds) in order to align it to the reference molecule. See example (B) in Figure: Example of the molecule alignment.
- suppressH
If true, than explicit hydrogens are suppressed in the input molecule by calling the
OESuppressNonDepictionHydrogensfunction.
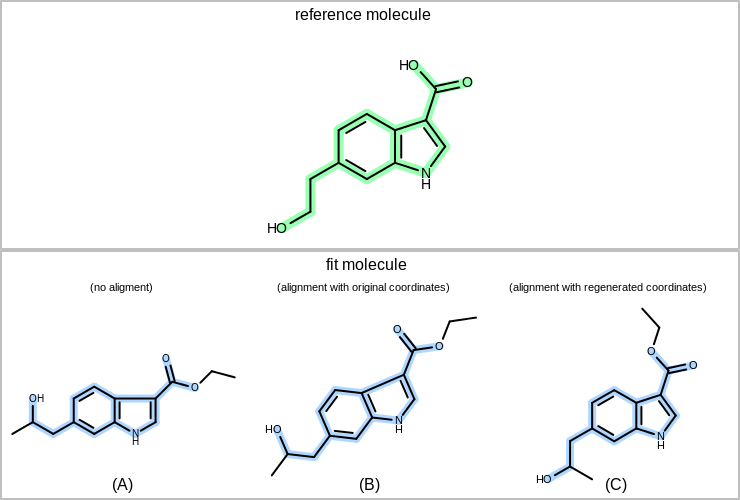
Example of the molecule alignment
(A) molecule with no alignment; (B) aligning the fit molecule to the reference molecule by allowing only 2D rotations (original coordinates are kept); (C) aligning the fit molecule to the reference by allowing rotation around single bonds (coordinates are regenerate)
See also
Molecule Alignment chapter
bool OEPrepareAlignedDepiction(OEChem::OEMolBase &fitmol,
const OEChem::OEMolBase &refmol,
const OEChem::OEMatchBase &match,
const OEAlignmentOptions &opts)
Generates the 2D coordinates of a molecule by maximizing its overlap with a reference 2D molecule based on the given match between the two molecules.
- fitmol
The molecule being fitted to be aligned to the reference molecule.
- refmol
The reference molecule of which 2D coordinates are remain unchanged.
- match
The match determines the atoms of the fitmol that will be aligned to the atoms of the refmol. The match can be generated by hand, or with any of OEChem TK‘s matching algorithms such as OESubSearch or OEMCSSearch.
- opts
The OEAlignmentOptions object that stores properties that influence how the molecule alignment is performed.
See also
Molecule Alignment chapter
OEAlignmentResult OEPrepareAlignedDepiction(OEChem::OEMolBase &fitmol,
const OEChem::OEMCSSearch &mcss,
const OEAlignmentOptions &opts=OEAlignmentOptions())
Generates the 2D coordinates of a molecule by maximizing its overlap based on the maximum common substructure initialized with a reference 2D molecule.
- fitmol
The molecule being fitted to be aligned to the reference molecule.
- mcss
The OEMCSSearch object stores the reference molecule. The molecule alignments are performed based on the maximum common substructure(s) detected between the fit and the reference molecule.
- opts
The OEAlignmentOptions object that stores properties that influence how the molecule alignment is performed.
Warning
The reference molecule has to have 2D coordinates prior to initializing the OEMCSSearch object.
See also
OEMCSSearch class in the OEChem TK manual
Listing 1example code in Molecule Alignment chapter
OEAlignmentResult OEPrepareAlignedDepiction(OEChem::OEMolBase &fitmol,
const OEChem::OESubSearch &subsrc,
const OEAlignmentOptions &opts=OEAlignmentOptions())
Generates the 2D coordinates of a molecule by maximizing its overlap based on substructure search initialized with a reference 2D molecule.
In case of a successful alignment, the returned OEAlignmentResult object stores the match between fit and reference molecules.
- fitmol
The molecule being fitted to be aligned to the reference molecule.
- subsrc
The OESubSearch object stores the reference molecule. The molecule alignments are performed based on the substructure(s) detected between the fit and the reference molecule.
- opts
The OEAlignmentOptions object that stores properties that influence how the molecule alignment is performed.
Warning
The reference molecule has to have 2D coordinates prior to initializing the OESubSearch object.
See also
OESubSearch class in the OEChem TK manual
Listing 2code example in Molecule Alignment chapter
OEAlignmentResult OEPrepareAlignedDepiction(OEChem::OEMolBase &fitmol,
const OEChem::OEMolBase &refmol,
OESystem::OEIter<const OEChem::OEMatchBase> &miter,
const OEAlignmentOptions &opts=OEAlignmentOptions())
OEAlignmentResult OEPrepareAlignedDepiction(OEChem::OEMolBase &fitmol,
const OEChem::OEMolBase &refmol,
OESystem::OEIterBase<const OEChem::OEMatchBase> *miter,
const OEAlignmentOptions &opts=OEAlignmentOptions())
Generates the 2D coordinates of a molecule by maximizing its overlap with a reference 2D molecule based on the given matches between the two molecules.
- fitmol
The molecule being fitted to be aligned to the reference molecule.
- refmol
The reference molecule of which 2D coordinates are remain unchanged.
- miter
The iterator that stores the matches on which the molecule alignment is based on.
- opts
The OEAlignmentOptions object that stores properties that influence how the molecule alignment is performed.
See also
Molecule Alignment chapter