2024.3.1 Release Notes
Highlights of the Current Release

We are pleased to offer many new features and improvements for Molecule Search. This tool is no longer a beta version.
You may opt in to run Orion jobs on the new Cadence Cloud for Molecular Sciences (CCMS), in addition to AWS. Contact us at sales@eyesopen.com for more information.
Molecule Search Query Sketcher
An upload query button in the toolbar now allows a user to upload a query from disk.
When a molecule with 3D information is uploaded to the Sketcher window, a 3D depiction will appear in the lower right side of the Sketcher window.
You can add another color representation to a current color atom by clicking the existing color atom. This is useful when working with ring color atoms, which are mapped relatively to multiple shape atoms. Previously, you had to Shift-click on each shape atom to add another color representation to a color atom that is mapped to multiple shape atoms. For more information, see the documentation about use of the color tool for 3D search.
Molecule Search Results Section
There is a progress bar for running FastROCS queries.
Project names are displayed on the Molecule Search History page. Under the Created column, the name of the the project is displayed that was in effect when the search was done. A slider switch controls whether only the current active project’s queries are shown.
A “copy” indicator appears when you hover on a Molecule Search result.
Figure 1. Copy button for a result tile.
When switching between 2D and 3D search result depiction types, the coordinates and rotations of molecules are retained.
In the 3D search results, the 2D and 3D scores have been added to the depiction panel that appears when hovering over a point in the plot.
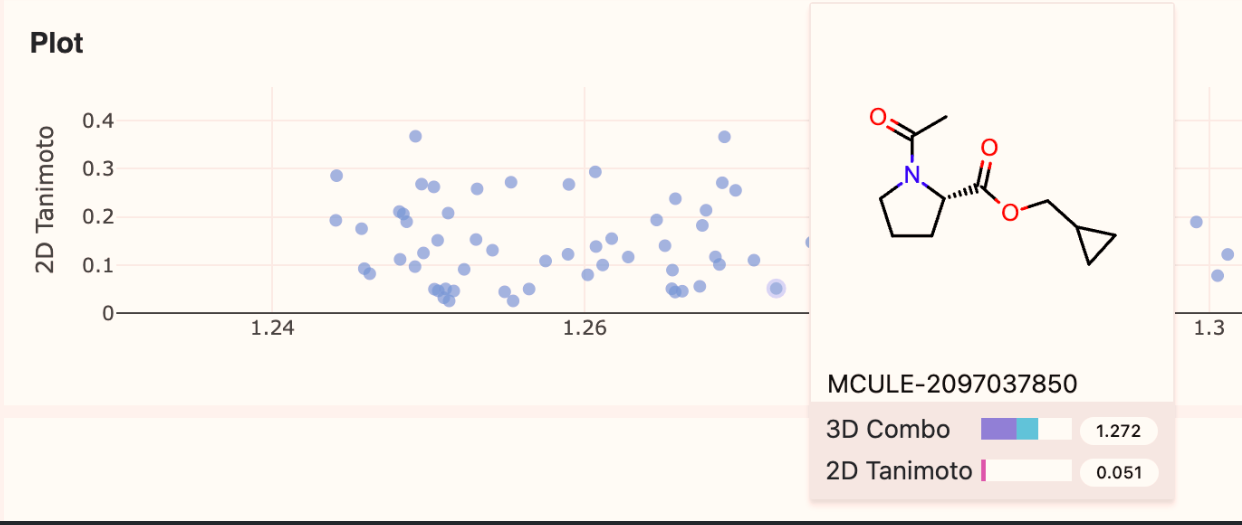
Figure 2. Scores displayed for a datum.
A ligand from a design unit on the 3D page can be sent directly to Molecule Search with a right-click gesture.
Similarity score labels have been improved for 3D similarity searches. When possible, the full terms are spelled out:
3D Tanimoto Combo
3D Shape Tanimoto
3D Color Tanimoto
2D Tanimoto
In small result cards, these may be abbreviated to “3D” and “2D” or, if there is more space, “3D Combo” and “2D Tanimoto.”
There is now an option to sort the 3D similarity search results by 2D Tanimoto scores.
User Account Roles for Molecule Search
A new standalone-search-user role provides a dedicated interface for users who only need Molecule Search. Contact us at sales@eyesopen.com for more information.
A new feature enables the assignment of a default role for any organization. The default role choices are: chemist, modeler, mt-modeler, and standalone-search-user.
A new search-admin role has been added for managing custom molecule search databases, but still only managed stack users can load their own custom databases.
Data Page
Deprecation of Search Services and Databases
There is a new policy for deprecating and removing old versions of OpenEye-provided FastROCS and Gigadock™ databases.
The legacy MaaS and FastROCS services will be disabled for SaaS customers and the Sources page will be replaced with a new Macro Molecules page that only includes MMDS.
Managed Service customers still using the legacy MaaS or FastROCS services are encouraged to migrate to Molecule Search. Databases provided by OpenEye are available in Molecule Search without the additional hosting costs that the legacy MaaS and FastROCS require.
Bug Fixes
In a Molecule Search query, you can highlight bonds even if an atom has not been selected.
Exact searches return only the requested maximum number of hits.
The protein component of a design unit from the LARGE MOLECULES or ALL DATA windows of the 3D page can be renamed.
A bug has been fixed that caused an invalid link to be formed from using the Copy Link to Folder Path icon (
 ).
).
Information for Programmers
Managed Service customers can develop their own cubes and floes for execution in Orion. They can also develop and manage molecule search databases. For more information, see the Programming Guide.