Design Decisions
This chapter seeks to give a general overview of the major design decisions behind OEChem TK. The reader should have an understanding of the basic OEChem TK objects and functions. The reader should also be familiar with the basics of object oriented programming.
Throughout this manual, the OEMol,
OEGraphMol, and OEQMol are the
concrete classes which handle most molecular representation in
OEChem TK. The OEGraphMol API is defined in the
OEMolBase abstract base-class, the
OEMol API is defined by the
OEMolBase and
OEMCMolBase abstract base-classes, and
the OEQMol API is defined by the
OEMolBase and OEQMolBase abstract
base-classes. The OEMCMolBase class
publicly inherits from the OEMolBase class, which in
turn inherits from the OEBase class. Similarly, the
OEQMolBase class publicly inherits from the
OEMolBase class, which again inherits from the
OEBase class. An OEGraphMol, can
be passed to any function which takes an OEMolBase
argument. An OEMol can be passed to any function
which takes an OEMCMolBase or
OEMolBase argument, and an OEQMol
can be passed to any function which takes an
OEQMolBase or OEMolBase
argument. Figure: Hierarchy A represents the OEChem TK
molecule hierarchy described in this paragraph.
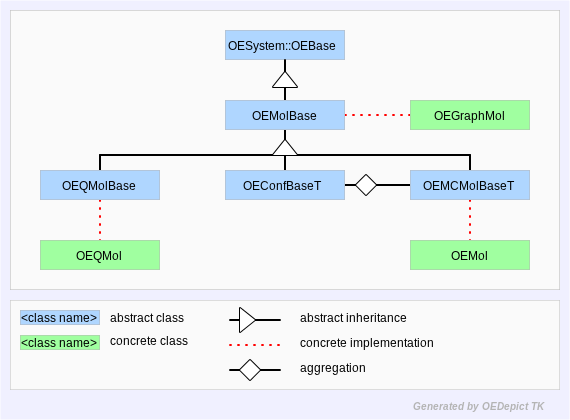
Hierarchy A
OEChem TK Molecule Hierarchy
Abstract Base-Classes
The most important data types in the OEChem TK library are OEMolBase, OEAtomBase, and OEBondBase. These three classes describe the behaviors of molecules, atoms, and bonds respectively. However, these types are abstract classes, describing the methods and semantics of molecules, atoms and bonds, but without defining an actual implementation. (See Figure: Hierarchy B)
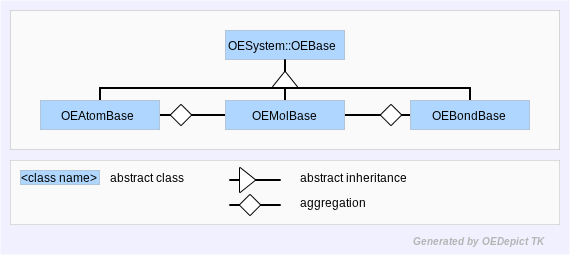
Hierarchy B
OEChem TK Abstract Base-Classes
Hiding the implementation of these types is very important to the longevity of OEChem TK. It avoids the problem of the leaky abstraction. If code was written that was expecting a certain behavior from the implementation it would be fragile to arbitrary changes in implementation. Therefore, these abstract base-classes are considered stable, with a guarantee that they will not change from version to version.
Note
Since OEAtomBase, OEBondBase, and
OEConfBase can only be accessed
through their parent molecules, there is no need for concrete
instances of these classes. In OEChem TK, these three classes are
accessed via references to their respective base-classes, or
through the iterator interface discussed in the
Atom and Bond Traversal chapter.
Factory Functions
It is standard practice when working with abstract base-classes, such as the OEMolBase, to define a function which returns a pointer to one of these objects. These functions, called factories, give library users access to concrete objects even when only the abstract base-class is exposed in the public API. The following factory functions are provided to create molecule objects in OEChem TK. The associated symbolic namespace is used to control what type of molecule implementation is returned.
Factory Function |
Symbolic Namespace |
|---|---|
Smart Pointers
The problem with factory functions is that they require the user to
manage the object’s memory. When the factory function returns a
pointer to an abstract base-class, it also passes ownership of the
memory to the programmer. To alleviate the problems associated with
memory management introduced by factories, the smart-pointer idiom is often
used. Simply put, a smart-pointer holds a real pointer to an object,
and deallocates the pointer’s memory when the smart-pointer goes out
of scope (e.g. in its destructor). In OEChem TK,
OEGraphMols,
OEMols, and OEQMols
fulfill the function of both factories and smart-pointers. This gives
the user access to multiple implementations without the need of
worrying about memory management. The constructors allow the user to
specify which implementation they would like using the associated
symbolic namespace described in the previous table. Then the objects
themselves act as smart-pointers, cleaning up the implementation
pointers when the molecules go out of scope.
Listing 1 demonstrates how to specify an
alternative molecule implementation. A SMILES
string is then read into different implementation. This
demonstrates how implementations that adhere to the
OEMolBase interface can reuse existing algorithms
written towards it.
Listing 1: Using an alternative molecule implementation
from openeye import oechem
mol = oechem.OEGraphMol(oechem.OEMolBaseType_OEMiniMol)
oechem.OESmilesToMol(mol, "C1=CC=CC=C1")
print("Canonical isomeric SMILES is", oechem.OEMolToSmiles(mol))
Query Molecules
It is not uncommon in chemical informatics to consider the equivalence of the graph which represents a molecule and the graph which represents a substructure query. Indeed the simplest of queries are molecules themselves. If one considers each node (or atom) as a potentially complex atom query, even highly complex queries can be represented as molecules.
In OEChem TK, this concept of a query as a molecule is represented by the OEQMolBase abstract base-class. An OEQMolBase contains OEQAtomBase and OEQBondBase atom and bond representations. These versions of OEChem TK atoms and bonds manage the atom and bond expressions which comprise the query.
OEChem TK defines the concrete OEQMol class which provides a programmer access to the API defined by the OEQMolBase class. This is analogous to the way an OEGraphMol provides concrete access to OEMolBase API. Figure: Hierarchy C represents the OEChem TK query molecule hierarchy.
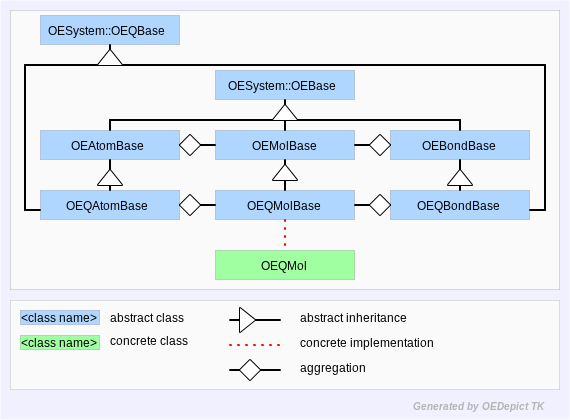
Hierarchy C
OEChem TK Query Molecule Hierarchy
This abstraction allows OEChem TK to treat separate query mechanisms to
work similarly. This is similar to how OEChem TK can represent any
molecular file format through a single OEMolBase
interface. For example, SMARTS can be parsed
into an OEQMolBase using the
OEParseSmarts function. Also, MDL Queries can be parsed into an
OEQMolBase using the
OEReadMDLQueryFile file.
See also
Pattern Matching for a general discussion of OEChem TK pattern matching facilities.
Free-Functions
OEChem TK is an object-oriented library. However, the design philosophy is that molecule objects are primarily data containers with data access member functions. Most powerful data analysis and manipulation routines in OEChem TK are implemented as free functions rather than member functions. This decision is based on the realization that the abstraction of a molecule can be neither stable nor consistent. To one programmer, a molecule should describe and perceive the space group of organometallic complexes, while to another a molecule should describe the residues and secondary structure of a macromolecule. Both of these perspectives are reasonable and should be supported. Further, a nearly infinite additional list of molecule designs can be imagined and should be supported. In order to do this, the OEChem TK molecule must be extensible, light-weight, and easily re-implemented. Thus, major changes to the OEChem TK molecules can be made, without need to re-implement more than a handful of functions. Conversely, an entire new area of chemistry can be added to the OEChem TK repertoire through free-functions without needing to implement the function in multiple molecules. We have used namespaces extensively to keep the free-functions from cluttering the global namespace.
See also
The OEChem Functions API section for a complete listing of all of OEChem TK’s free-functions.
Programming Layers: The Deep and Twisted Path
OEChem TK was designed to provide a library which puts powerful algorithms in the hands of novice users without hand-cuffing the expert. For this reason, OEChem TK can at the same time seem trivial and overwhelming. There are often several ways to carry out certain tasks in OEChem TK each with its subtle advantages, which can benefit an experienced user. There are very few algorithms we have shied away from including in OEChem TK, and many of the methods are new, unique and powerful. This gives OEChem TK a very rich interface, yet to gain this efficiency and power OEChem TK may force the user to think about problems in ways they are not accustomed to doing. The hope is that the user can benefit from the experience.
OEChem TK has several layers of interfaces to most of its
functionality. There are “high-level” interfaces, which provide the
user with an enormous amount of power with minimal code. This level is
exemplified by the OEReadMolecule and
OEWriteMolecule functions. With these functions the
functionality of the babel file-format conversion program is
trivial. While this is trivial to write and understand (maybe after
understanding this manual), it should not belie the fact that OEChem TK
is carrying out an enormous amount of work under the surface.
A perhaps “mid-level” interface in OEChem TK is the ability to fine tune molecule file-formats using flavors described in Flavored Input and Output section. Flavors are most useful when file-formats are being used for more than they were originally intended, for example, PDB files. While this functionality is perhaps not for the first-day user, it certainly doesn’t require a stout heart.
Finally, for advanced programmers, OEChem TK provides access to nearly
all of the details. OEChem TK molecules have a simple API which can be
used to derived custom molecule implementations. The free-function
heavy API lets the user apply OEChem TK’s powerful algorithms on custom
data structures. Similarly, many of the functions that are wrapped in
high level functions (like the molecule readers and writers) are also
available directly to the user at the low level. For instance,
OEWritePDBFile allows the user to write out a PDB
file with a very specific flavor without any normalization that
OEWriteMolecule may perform.
OEChem TK is a still a live moving software project. If a roadblock is found a particular level, dig deeper into the next level. Often, the functionality is already present.