OEMolFunc
Attention
This API is currently available in C++ and Python.
class OEMolFunc
The OEMolFunc is an abstract base class. This class defines the interface for functions that require a molecule to compute the function value, and is often associated with a force field. When associated with a force field, this represents a forcefield component or a collection of components with each component being a collection of interactions (OEInteraction). Thus, classes derived from OEMolFunc can represent a part of or a complete force field. The force field components need to be done concomitant with one or more OEFFParams derived classes.
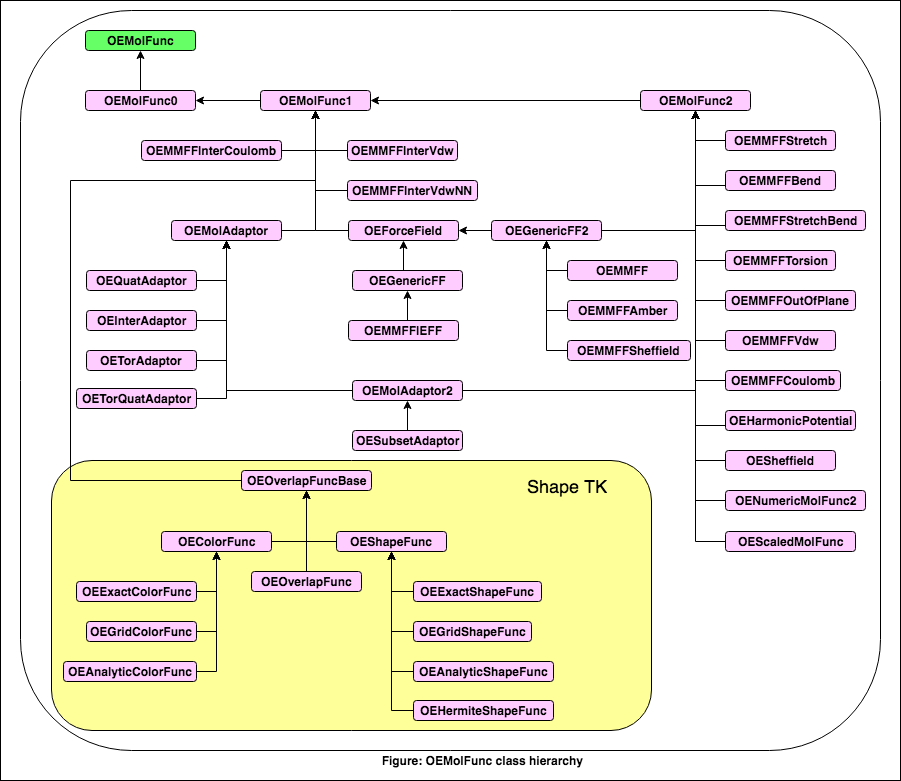
- The OEMolFunc class defines the following public methods:
- The following classes derive from this class:
Clear
void Clear()
Deletes all interactions contained in the OEMolFunc derived instance.
The interactions are created during setup (OEMolFunc.Setup).
ClearPredicates
void ClearPredicates()
Deletes all stored predicates set, previously created by using one of the overloaded
OEMolFunc.Set methods. Predicates control function behavior
during setup (OEMolFunc.Setup).
GetInteractions
OESystem::OEIterBase<OEInteraction> *GetInteractions(const OEChem::OEMolBase &,
const double *,
unsigned functype=0) const
This method defines the interface for retrieving interaction level information computed by the OEMolFunc instance, given a molecule (first argument) and a set of coordinates (second argument). The method returns an iterator over the OEInteraction objects. See the section on OEInteraction for the details regarding interaction interrogation and reporting.
If OEMolFunc is a composition of multiple components,
the last argument may be used to request the interactions of only a single component by passing a value from the OEFuncType namespace.
Due to the unique construct of the return value from this method, it is not possible to implement this in a user derived class. Default implementation of this method return an iterator over an empty vector.
Set
bool Set(const OESystem::OEUnaryPredicate<OEChem::OEBondBase> &pred,
unsigned int functype)
bool Set(const OESystem::OEUnaryPredicate<OEChem::OEAtomBase> &pred,
unsigned int functype)
bool Set(const OESystem::OEBinaryPredicate<OEChem::OEAtomBase,
OEChem::OEAtomBase> &pred, unsigned int functype)
bool Set(const OESystem::OEBinaryPredicate<OEChem::OEBondBase,
OEChem::OEBondBase> &pred, unsigned int functype)
These methods assigns interaction-control predicates to the OEMolFunc
derived instance. The first argument is the predicate type to be passed to the interaction component.
The second argument is a value or a set of binary OR’ed values taken from the OEFuncType
namespace which specifies the intended target for the predicate assignment.
Most derived classes can only use a handful of the four different kinds of predicates
allowed in these overloads, thus only the specific overloads are overwritten by various derived class.
The default implementation for all the overloads has trivial implementation and returns a false.
Setup
bool Setup(const OEChem::OEMolBase&)
This method defines the interface for initializing a OEMolFunc derived instance with a molecule. All classes derived from OEMolFunc implements this method. Typically the implementation of this method requires perception of the passed molecule for efficient energy evaluation.
See also
SetupConf
bool SetupConf(const double *)
This method defines the interface for initializing a OEMolFunc derived
instance with coordinates of a new conformer, after it has already been initialized with the molecule by
calling Setup. Not all classes derived from
OEMolFunc implements this method, but calling this should never
cause any additional performance issue or failures.