How to Prepare Molecule Search Databases from Molecule Files
Preparing a molecule search database from a molecule file is a multistep process involving several floes, where the outputs of earlier floes are passed in as inputs to later floes. These intermediate resources are collections, some of which can be used as input to other floes, such as FastROCS.
Note
For assistance in creating a custom database, SaaS and Standalone Search admins should first contact OpenEye Customer Support at support@eyesopen.com.
Please see the Custom Databases for Molecule Search section for additional details about this process.
The Automated Preparation for Molecule Search Databases Floe supports these common molecule file formats as input: .smi, .ism, .cxsmiles, .csv, and .sdf. This lowers the barrier to creating custom molecule search databases by providing a convenient, end-to-end automated workflow.
This orchestration floe automates the database preparation process but could potentially stop at any intermediate stage in the process. The term child floe will be used here to refer to floes launched by this floe.
The 2D cube group is the series of three steps that converts a molecule file into a 2D molecule search database. It corresponds to the three cubes in the yellow box in Figure 1. The first two cubes each correspond to a child floe running in the background, and the third cube runs code that builds the database. The 3D cube group is the series of three steps that converts a molecule file into a 3D molecule search database. It corresponds to the three cubes in the red box below. As with the 2D cube group, the first two cubes each correspond to a child floe running in the background, and the third cube runs code that builds the database.

Figure 1. The floe map of the Automated Preparation for Molecule Search Databases Floe. The yellow box shows the 2D cube group, and the red box shows the 3D cube group.
Provide the Parameters
Navigate to the Workfloes tab on the Floe page and select the Molecule Search Floes package in the left pane. Select the Automated Preparation for Molecule Search Databases Floe and click “Launch Floe” to bring up the Job Form.
The Job Form contains the parameters needed to run the floe and can be modified as desired.
Prep Input
Input File: Click the “Choose Input” button to bring up a list of your existing files and select the input file you want to use. The file must be in .smi, .ism, .cxsmiles, .csv, or .sdf format. When you upload your file, be sure that the “Convert to datasets” box is unchecked.
Note
Recommended size limits are approximately 900M molecules for 2D and 200M molecules (2B conformers) for 3D. Preparing databases with collections larger than these recommended limits should only be done after consulting with OpenEye support.
Identifier Strings
The name and version of your databases are specified in this section. The name of your database will be a composite name of the vendor library name and vendor version, separated by a space.
Custom Library Name: This is the name of the custom library, for example, Tutorial Database.
Custom Library Version: This is the database version of the custom library, to distinguish between the original and updated databases. We recommend using 1.0 as the number for the first version of a new database.
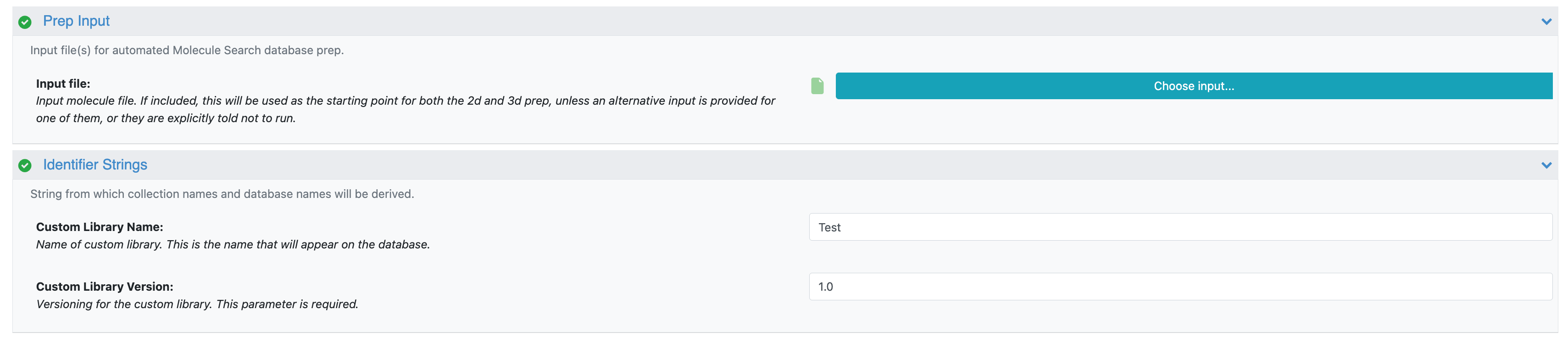
Figure 2. The Job Form showing the Input and Identifier Strings parameters.
Prep Automation Options
This section contains various parameters to control the behavior of the database preparation.
Ending Points: These parameters allow you to direct the floe to terminate the process prior to creation of the molecule search database, at the chosen intermediate stage.
Execute 2D/3D Prep: These parameters can be used to toggle the 2D or 3D branch, if you would rather generate only a single database type (2D or 3D) rather than both.
Keep DBs Loaded: Databases are UNLOADED by default upon completion of this floe but can be switched to LOADED using the toggle.
Note
Molecule search databases are only searchable when they are in the LOADED state. A LOADED database has a compute resource allocated and is loaded in memory and searchable. The LOADED state continuously accrues costs.
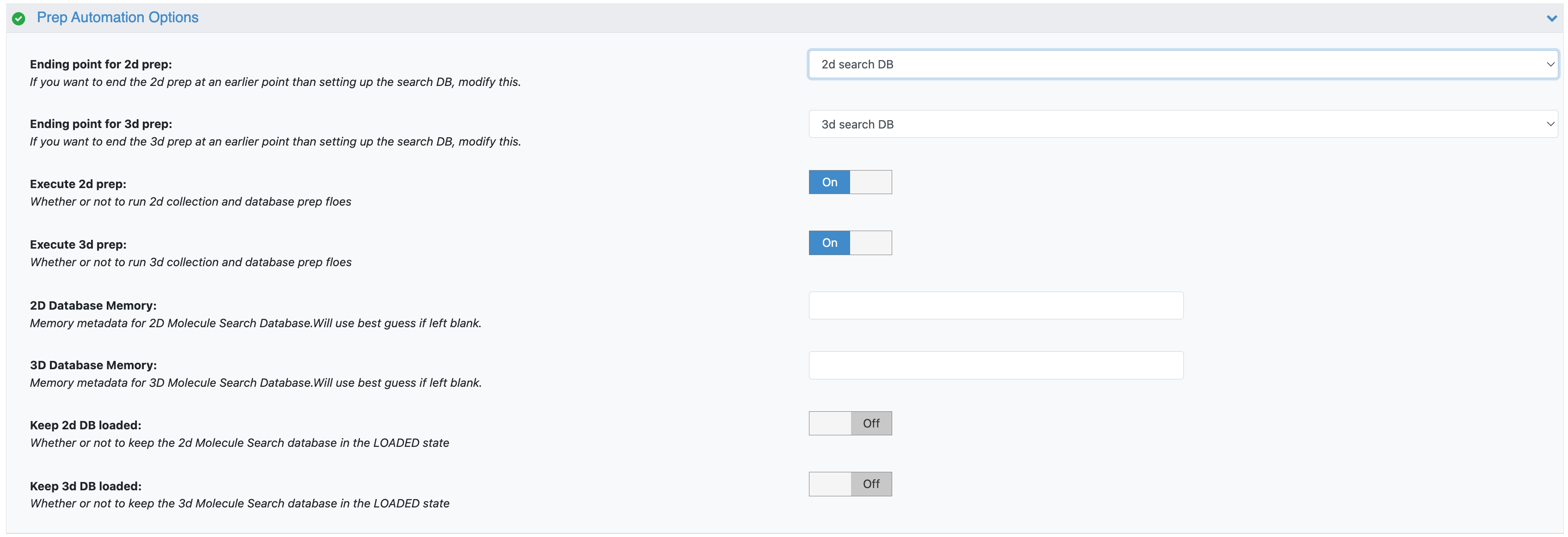
Figure 3. The parameters under the Prep Automation Options.
GigaPrep Parameter Group
The Prepare Giga Collections Floe is the child floe that prepares a FastROCS collection that is used as input for the 3D Molecule Search Database (triggered by the first cube in the 3D cube group in Figure 1).
Molecule Title: This parameter should be populated when your input file is in CSV format and you need to specify the title of the column containing the molecule titles.
OEFilter Type: BlockBuster is the default filter. Other filters may be selected from the drop-down menu. This is in keeping with the default parameters of the Prepare Giga Collections Floe.
The remaining GigaPrep Parameters are parameters that have been modified from the child floe’s defaults. See the GigaPrep tutorial for more information.
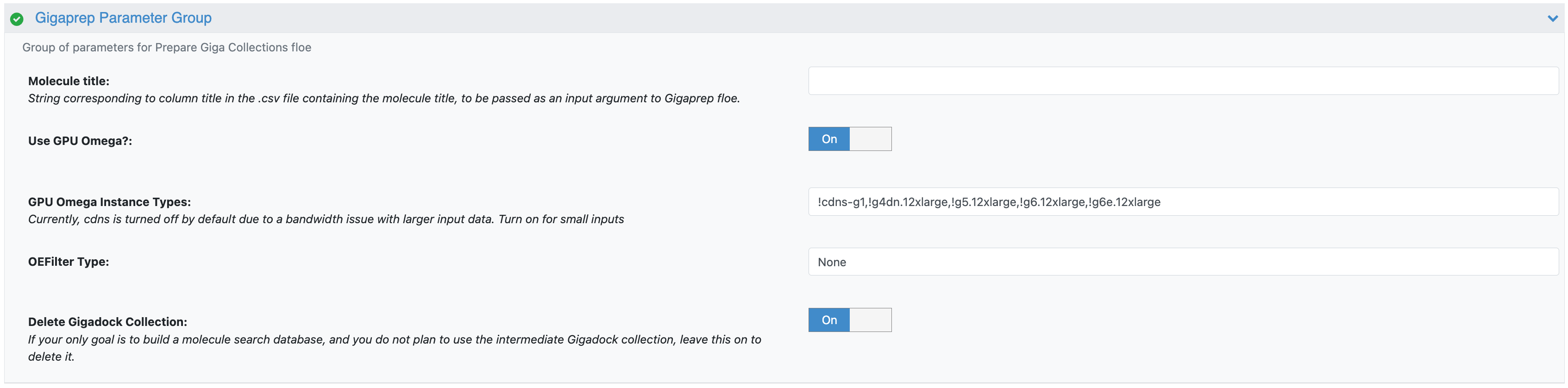
Figure 4. The parameters for the Prepare Giga Collections Floe.
Run the Floe
When the parameters have been specified, click “Start Job” to begin the floe.
The automated floe will begin, then the child floes will start as the requisite cubes are reached.
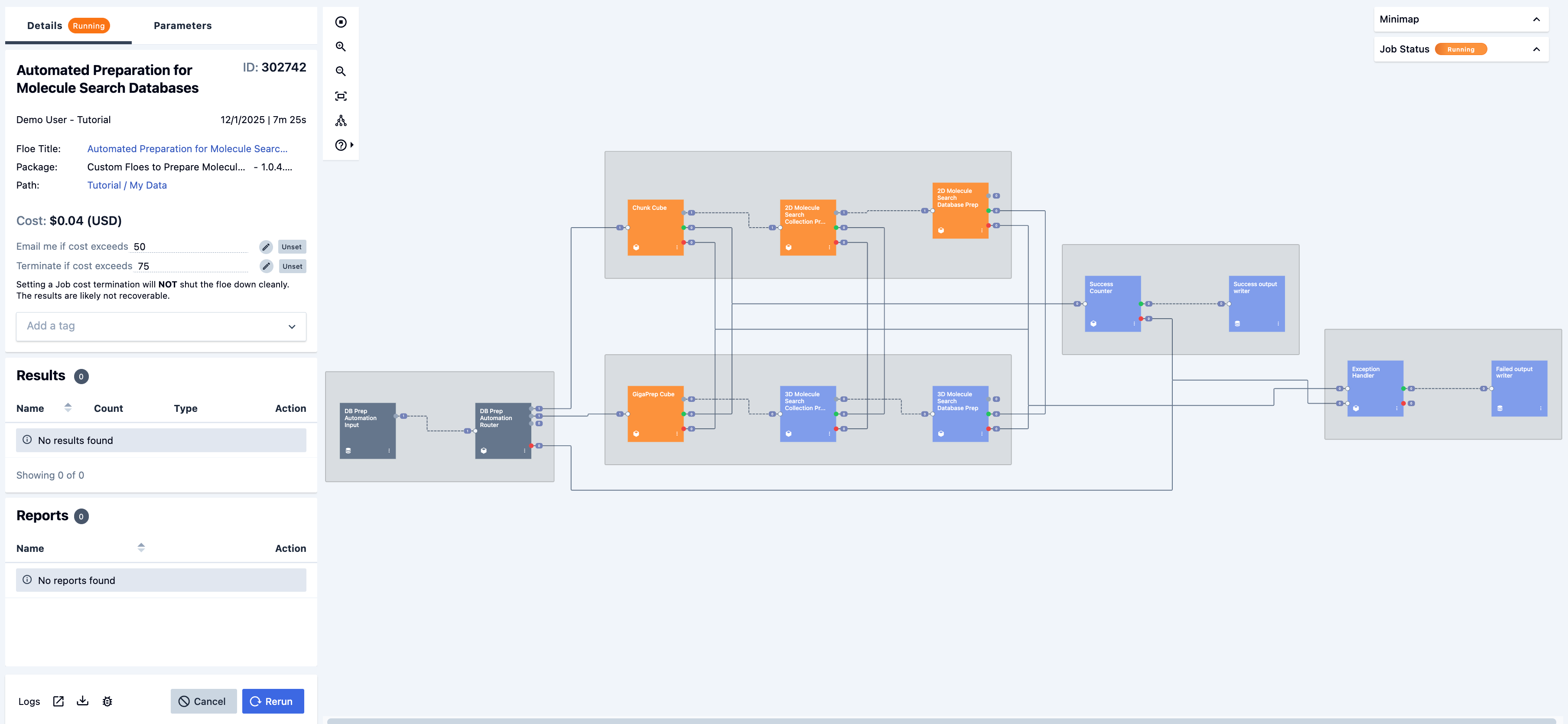
Figure 5. The running job for the Automated Preparation for Molecule Search Databases Floe.
The 2D Molecule Search Chunk Collection Prep Floe and the 3D Molecule Search GigaPrep Floe (seen in Figure 6) begin to run shortly after.

Figure 6. The running job for the 3D Molecule Search GigaPrep Floe.
The 2D Molecule Search Database Prep Floe begins next, as shown in Figure 7.
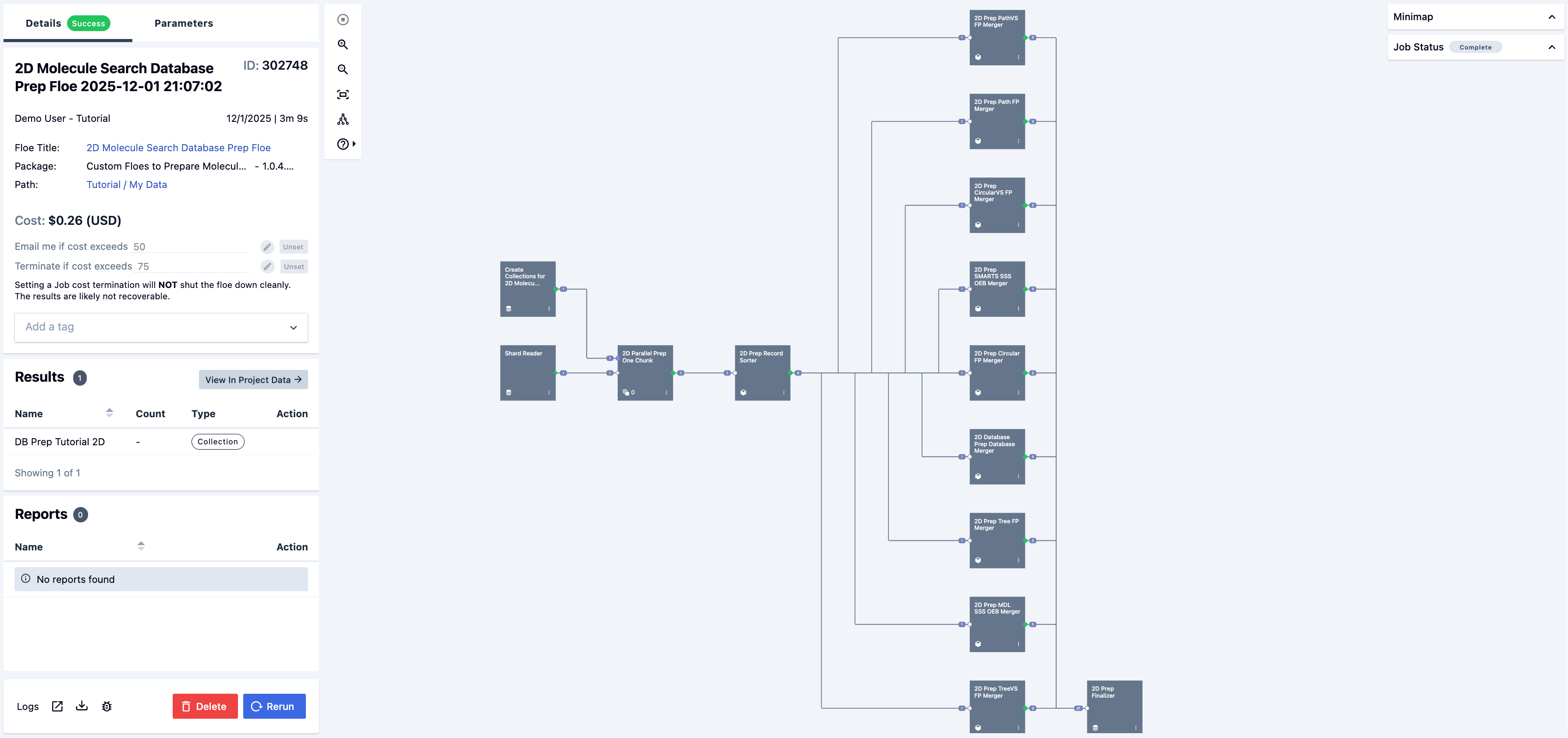
Figure 7. The completed job for the 2D Molecule Search Database Prep Floe.
The final child floe is the 3D Molecule Search Database Prep Floe, which begins as soon as the 3D Molecule Search GigaPrep Floe finishes and can be seen in Figure 8.
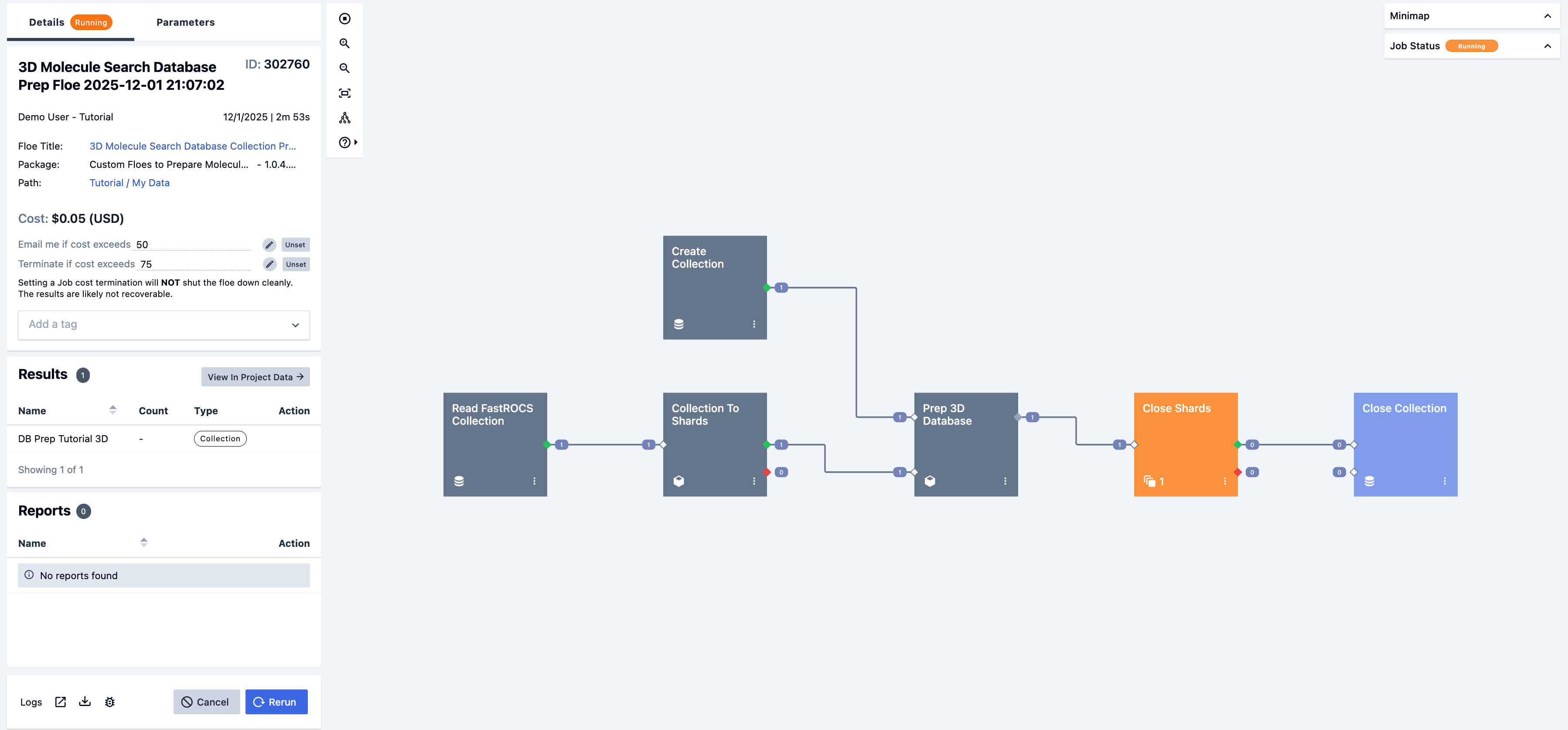
Figure 8. The running job for the 3D Molecule Search Database Prep Floe.
The Automated Preparation for Molecule Search Databases Floe will continue to run until the 2D and 3D databases have been built.
When the job is complete, the Automated Preparation Floe and its child floes will appear in the job list, as shown in Figure 9. The floe tags all of its child floes with its Job ID.
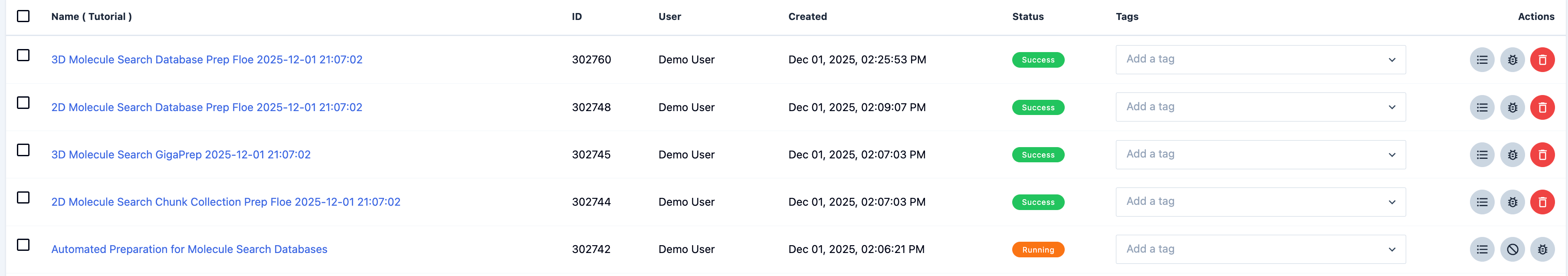
Figure 9. The job list showing the Automated Preparation for Molecule Search Databases Floe and its child floes.
Note
The 2D and 3D database tiles on the Databases tab in System will show the “QUEUED” status while the databases are being generated. This is normal behavior.
Expected Results
By default, the preparation floe produces a 2D and/or 3D database as the output. After a database is prepared, it will appear automatically as a custom database option on the Database Tab of the System page. At this point, the admin or modeler who created the database can load it for use in molecule searches.
Alternatively, if you opted to end the 2D and/or 3D preparation before setting up the search database (in the Ending Points parameters), then 2D and/or 3D search collections, or FastROCS search collections (for 3D), will be output as intermediate steps. These collections can be used as input for other operations. You can also upload other collections to your files. They will appear in the list of available collections when you click the “Create Database” tile on the Databases tab of the System page.
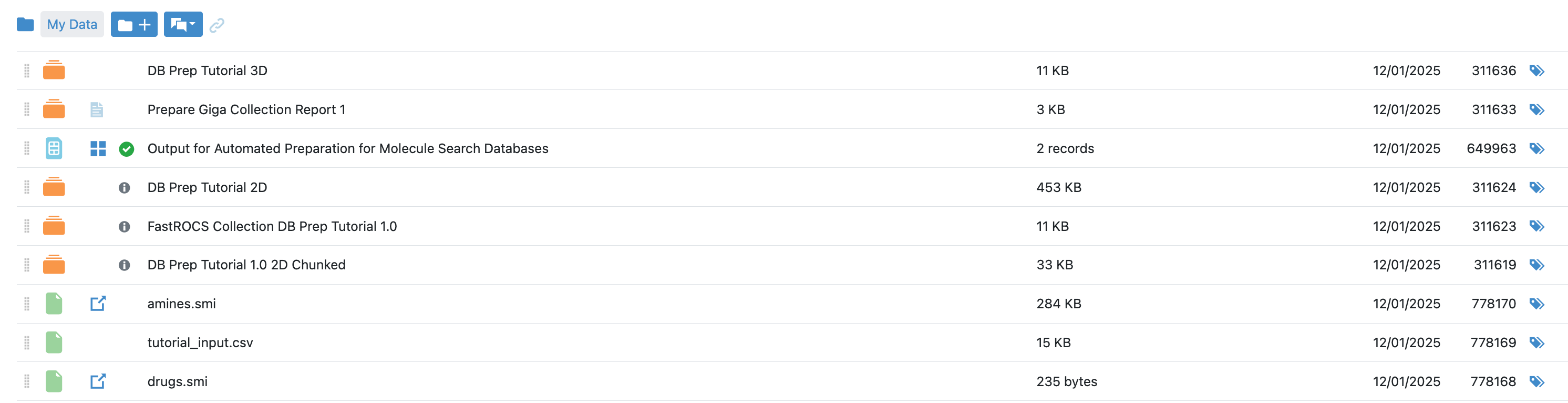
Figure 10. The list of databases created by the Automated Preparation for Molecule Search Databases Floe.
Approximate Costs
2D molecules (million) |
3D molecules (million) |
2D Cost |
3D Cost |
Total Cost |
|---|---|---|---|---|
2 |
4 |
$15 |
$25 |
$40 |
5 |
7 |
$15 |
$30 |
$45 |
23 |
27 |
$105 |
$795 |
$900 |
The floe can take a few hours for small (~1M molecules) databases. That increases drastically for larger databases due to 3D preparation, but it should complete within 24 hours.
Note
We recommend having at least 744 GB of memory and 6.3 TB of disk space available to prepare a 3D collection. These values are defined in the Database Prep Cube Instance Type parameter group in the 3D Molecule Search Database Prep Floe. They are defaulted to use larger instances to ensure enough resources are available to create databases. You may adjust for smaller instances after getting a sense of what is needed to make a database of a given size.