Version 2.2.0
OEChem TK 2.2.0
New features
Atom and bond expressions of query molecules have been optimized for substructure search. The graphs below show the performance improvement since the 2018.Oct release. Optimization has been turned on for both MDL queries and SMARTS patterns and can be controlled by the following options:
OESmartsParseOpts.Optimizewhen using theOEParseSmartsfunctionOEMDLQueryOpts.Optimizewhen using theOEReadMDLQueryFilefunction
Substructure search performance improvement for MDL queries 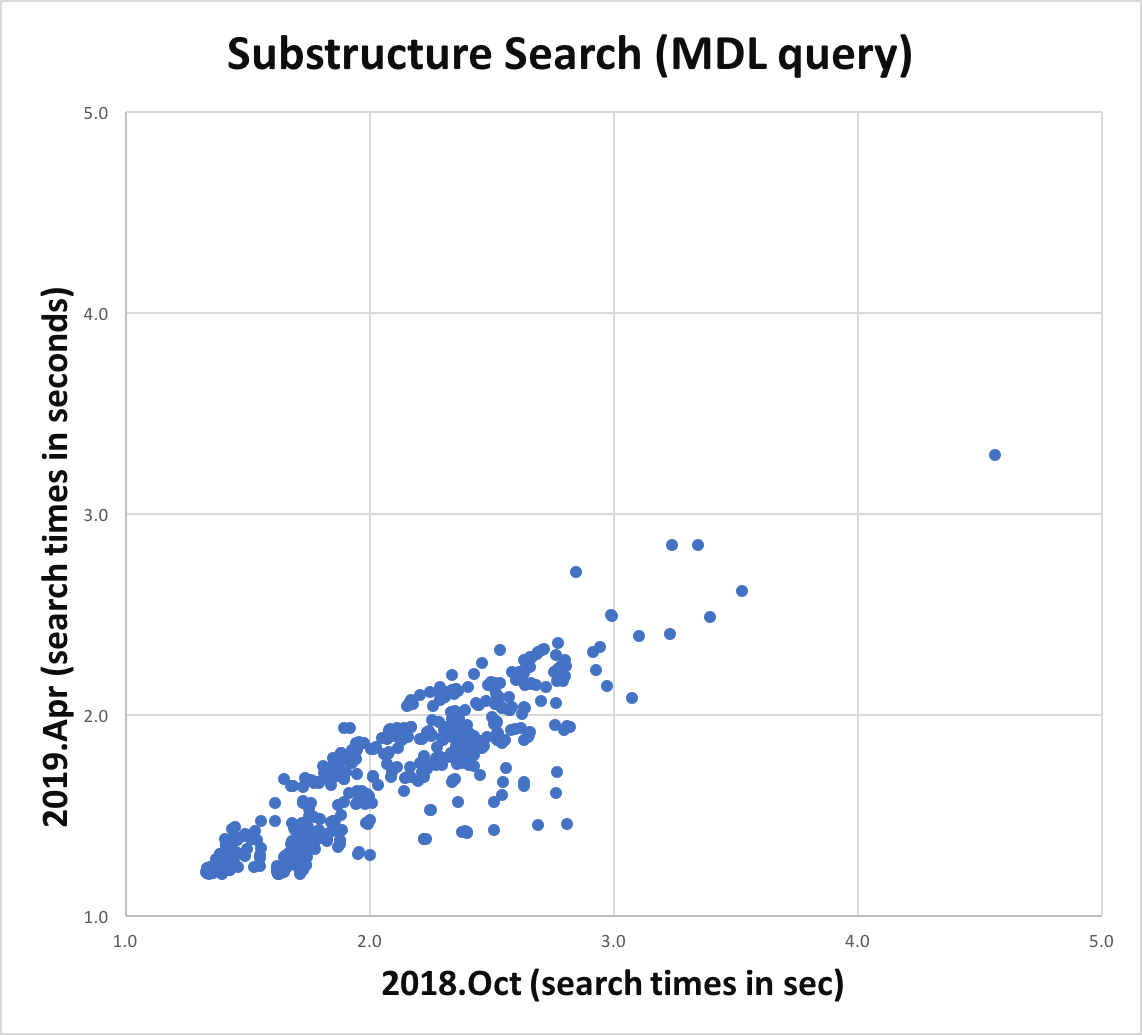
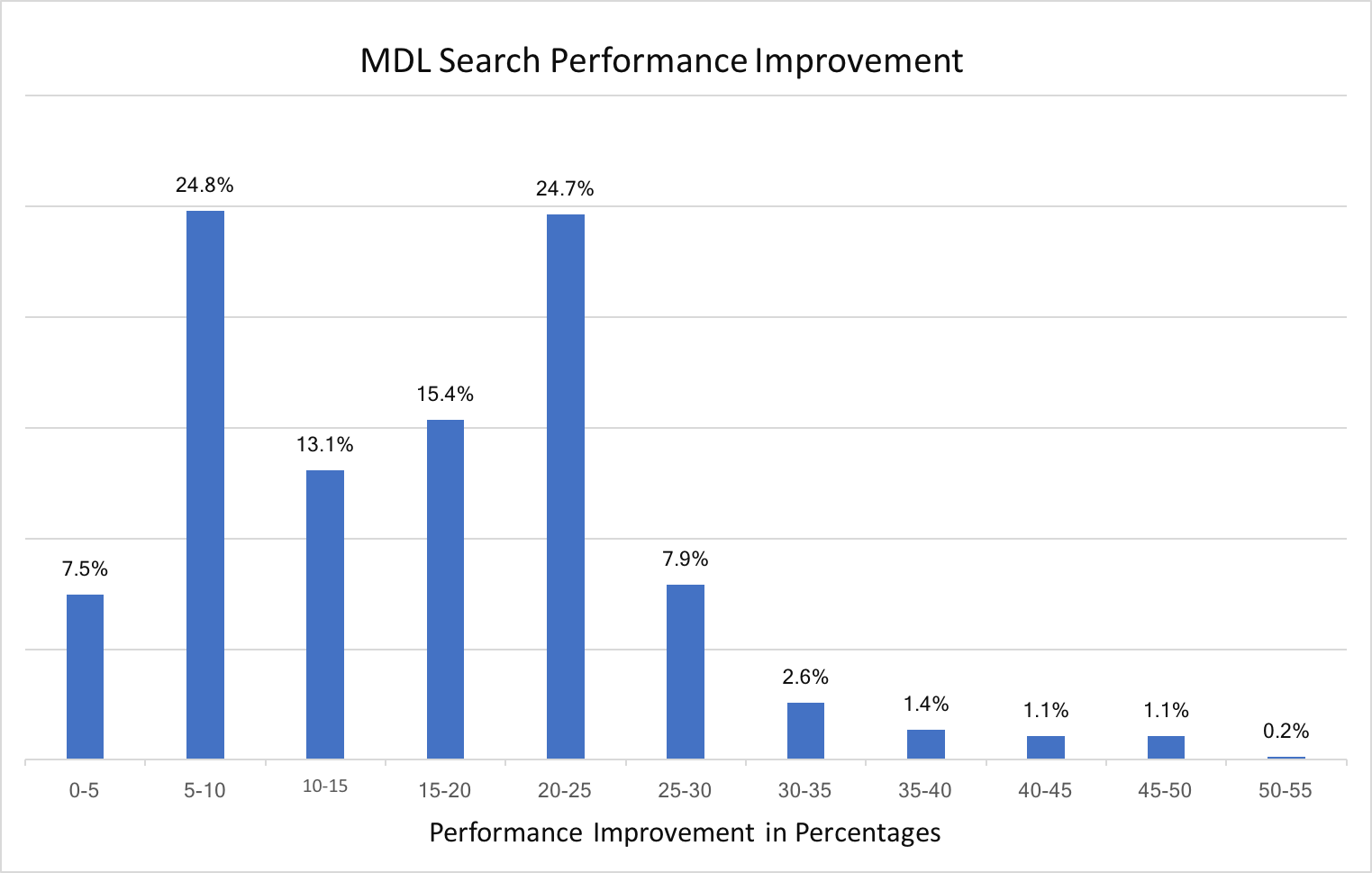
Substructure search performance improvement for SMARTS patterns 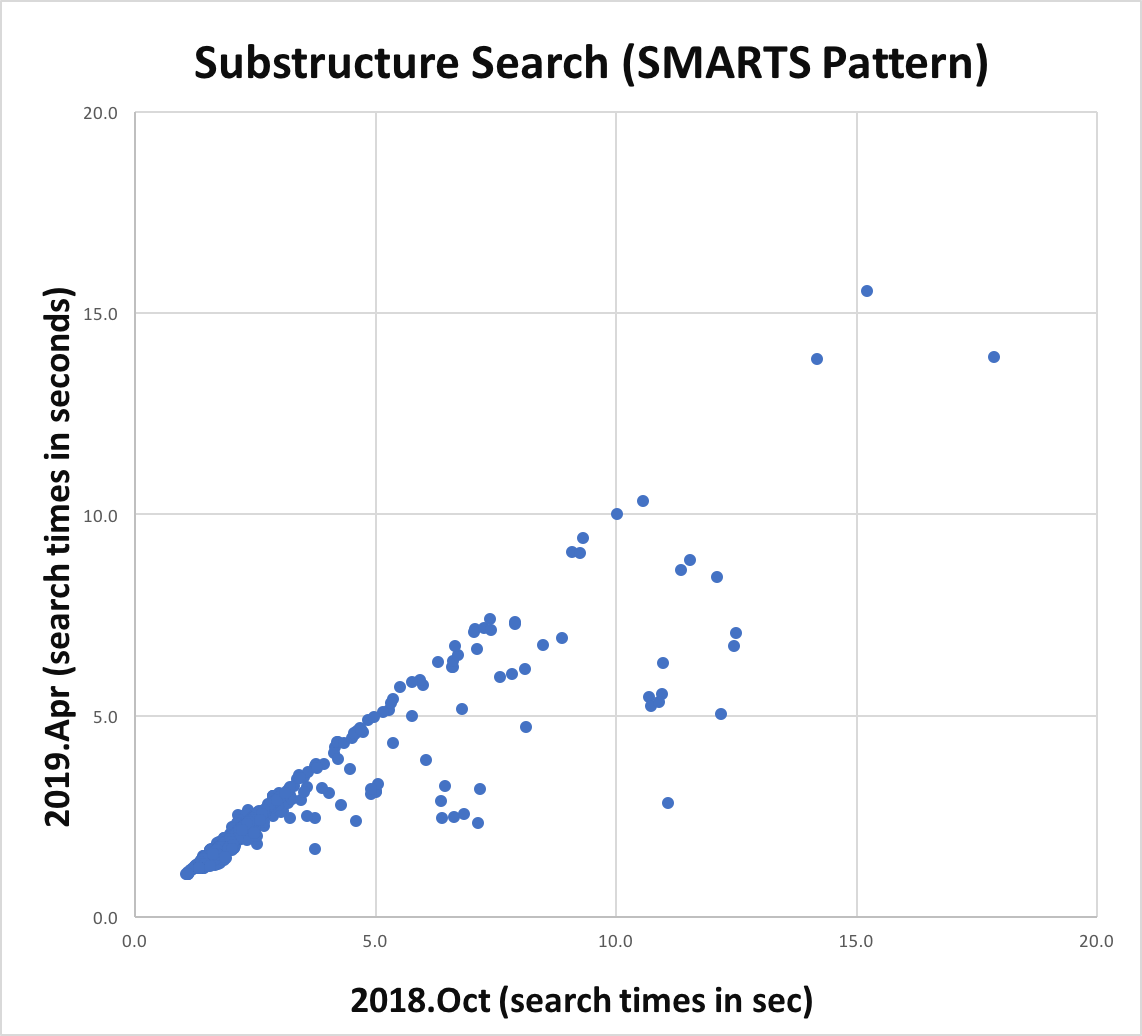
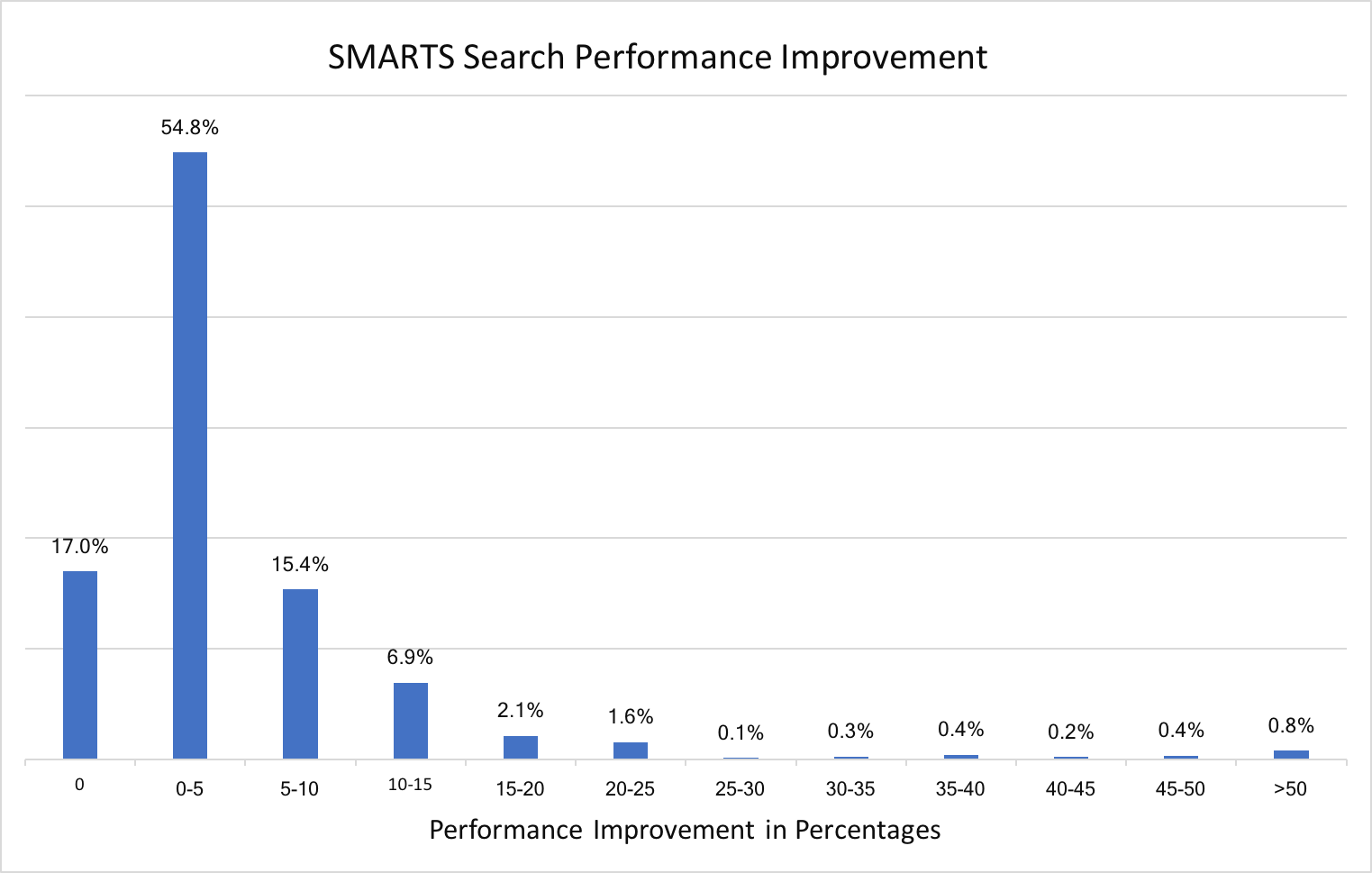
A new
OEFormat.OEZcompressed binary format has been added with the.oezextension. This file format only supports multi-conformer molecules derived from the OEMCMolBase class. Each molecule is individually compressed in the file using the Zstandard compression algorithm. See the Compressed Input and Output section for more details.Note
This new file format is not backwards compatible. OpenEye applications and toolkits released before April 2019 will not be able to read or write this file format.
An OEBitVector object now can be attached as generic data to any OEBase object. Its derived classes now round-trip through the OEB format.
Multi-conformer molecule overloads have been added to the following existing APIs:
New input flavors,
OEIFlavor.MDL.SuppressEmptyMolSkip,OEIFlavor.SDF.SuppressEmptyMolSkip, andOEIFlavor.RDF.SuppressEmptyMolSkip, have been added to the MDL formats to allow returning empty molecule records. This allows the SD data to be recovered from these records.New output flavors,
OEOFlavor.MDL.SuppressTimestampsandOEOFlavor.SDF.SuppressTimestamps, have been added to the MDL formats to explicitly suppress timestamps in the file header. This is convenient for generating diffable test files.A new
OEResidueToStringfunction has been added to convert an OEResidue object into string representation.The following new classes have been added to provide alternative ways to access tagged property data in molecules:
New predicates, OEIsValidAtomValence and OEIsValidMDLAtomValence, have been added to provide the ability to loop over atoms that satisfy the OpenEye and MDL valence model requirements, respectively.
The
OEMolBase.NewGroupmethod now also takes an OEAtomBondSet object.New
OECheckAtomValenceandOECheckAtomValencesfunctions have been added that check if an atom or all atoms of a molecule have valid valences.A new
OELibraryGen.NumPossibleProductsmethod has been added that returns the number of possible products that can be generated.New
OEClearSDDataandOEClearPDBDatafunction overloads have been added to clear SD or PDB data of multi-conformer molecules.A new
OEShortestPathoverload function has been added that disallows traversing excluded bonds in the path.A new
OEAtomBondSet.GetParentmethod has been added.A new
OEGetResidueIndexFromCodefunction has been added.The
OEPDBOrderAtomsfunction has been updated to take an argument that preserves the PDB residue order. It defaults tofalseto preserve backwards compatibility.
Major bug fixes
The number of cases where round-tripping to MDL file format results in stereo information loss has been significantly reduced.
Minor bug fixes
High-level SMILES readers such as
OESmilesToMolandOEReadMoleculenow clear cis/trans bond specifications on bonds in small rings (less than 7 members).The
oemolistream.openandoemolostream.openmethods now correctly set the formats forgzfilenames.A bug that caused a crash when an invalid oemolostream is finalized has been fixed.
oemolistream.Setgzandoemolostream.Setgzmethods are now a no-op if the state of the stream is unchanged.An issue that caused the JSON writer to crash with user-specified cis/trans stereo has been fixed.
The MDL V3000 reader has been fixed and no longer marks “either” double bonds with cis/trans parity.
The MDL V2000 reader has been fixed and no longer adds “either” double marks to chiral but unspecified cis/trans double bonds.
OESubsetMolno longer crashes when breaking and adding multiple R-groups to the same atoms.Cobalt ions are now assigned a formal charge when being read from the PDB file.
Python-specific changes
The
__str__method of the OEResidue class now invokes the newOEResidueToStringfunction.The
OEMolDatabase.GetMoleculeStringmethod has been fixed and now returnsbytes.
Documentation changes
New code snippets have been added to the
Substructure Searchsection to show how to reconstruct matches for the original molecule.A new Split molecule file example has been added.
New code snippets have been added to the
OEChemIsLicensedfunction.
OEBio TK 2.2.0
New features
A new
OEIsCTerminalAtompredicate has been added to identify the C-terminal atom(s) in a protein structure.A new
OEIsNTerminalAtompredicate has been added to identify the N-terminal atom(s) in a protein structure.A new OEAtomMatchResidue predicate has been added to identify residue atoms by RegEx.
New
OEGetResidueAtomfunctions have been added to provide easy access for specific residue atoms.The
OEIsBackboneAtompredicate has been updated to take an optional argument to consider the terminal oxygen atom (OXT) as part of the backbone.The
OEIsWaterpredicate has been updated to take two optional arguments:to match hydrogen atoms in water molecules
to match the water ions \(OH^{-}\) and \(H_{3}O^{+}\)
A new
OEIsStandardProteinResidueoverload that takes an atom has been added.
New features (Preliminary)
The following preliminary APIs have been added to handle design units:
The above classes and functions are currently used by Spruce TK for protein handling.
Minor bug fixes
The
OEPlaceHydrogensfunction has been improved:The
OEPlaceHydrogensMoverClass.NH2mover perception has been updated.The
OEPlaceHydrogensMoverClass.COOHmover perception has been updated to only mark the atoms involved in the mover.
Java-specific changes
The Java example in the Splitting a Macro-molecular Complex Efficiently and Flexibly section has been fixed.
Documentation changes
A new Interaction Perception chapter has been added.
Code snippets have been added to the
OESetCrystalSymmetryandOEGetCrystalSymmetryfunctions.
OESystem TK 2.2.0
Minor bug fixes
The
OEMakeMolecularGaussianGridfunction now returnsfalsefor an empty molecule.New code snippets have been added to the
OEThrowclass to demonstrate how to redirect warning and error messages to a file.
OEPlatform TK 2.2.0
New features
The following APIs have been rewritten using standard C++11 classes:
OEMutex now uses std::mutex.
OETryMutex now uses std::try_lock.
OELock and OELockSpinlock now use std::lock_guard.
Minor bug fixes
The
OEAddLicenseFromHttpfunction available in the wrapped languages (Python, Java, and C#) now accepts HTTP responses with atext/plain; charset=utf-8content type.
OEGrid TK 1.6.3
Minor bug fixes
The
OEReadMTZfunction has been updated to read CCP4 alternative space group numbers (e.g., 4005).OEReadGridno longer loses its title beyond the first blank space.A potential memory corruption issue with Gaussian volume calculation has been fixed.