Tutorial 1: NGS Pipeline with Custom Interactive Selection (PacBio), In-Vitro Library
Background
A selection campaign using a semi-synthetic, codon-optimized antibody library was carried out against the SARS-COV-2 antigen Spike (S) trimer, its subunit S1, and the S1 subunit RBD. Three independent selection campaigns were carried out using the same input library. DNA was isolated from a less stringent population, two rounds of in-vitro selection at 10 nM, and a more stringent population, three rounds of in vitro selection at 1 nM. Each population was prepared by PCR amplification with the addition of inline barcode consisting of 8mer (DNA) barcodes at the 5’ and 3’ ends to distinguish the six populations consisting of:
1) 2 x trimer (early and late rounds)2) 2 x S1 (early and late rounds)3) 2 x RBD (early and late rounds)
All input files can be located in Orion in following location:
Organization Data / OpenEye Data / Tutorial Data / AbXtract /
A barcode table titled ‘barcode_file_abxtract.xlsx’ indicates how these samples were prepared. Amplified DNA products were pooled together in a single tube and shipped to a PacBio / Illumina sequencing provider. The provider then shipped us back one PacBio file titled “pacbio_small_codon_optimized.fastq” and two Illumina files titled “F_illumina_codon_optimized_abxtract.fastq” and “R_illumina_codon_optimized_abxtract.fastq.” The PacBio file consists of a set of sequences each of length ~800 bp, composed of variable light (VL) and variable heavy (VH) sequences in VL-linker-VH orientation. The Illumina files consist of one forward and one reverse FASTQ file. Each sequence within the FASTQ is ~300 bp in length, with an overlapping region between the forward and reverse reads to enable paired end assembly of the sequences into one VH sequence.
For the purposes of this tutorial, each of the FASTQ files from PacBio/Illumina was reduced by taking a random output of 87,966 sequences for PacBio and 84,988 sequences from Illumina. The larger files from which these were derived contain an underlying diversity of full-length nucleotide of ~50K total, with a 20-fold reduction in the H3 diversity.
All populations are derived from the same input library against three overlapping subunits of the Spike (S) protein. The goal of this tutorial is to cluster all three populations binding to the Spike (S) trimer, which encompasses S1. S1 can further be broken into smaller units, one of which is the receptor binding domain (RBD). Because each domain is a subunit of the other, we anticipate overlap among RBD, S1, and the trimer, particularly since the same input library was used during this selection. We anticipate we will also find antibodies that are unique to select subunits (e.g., S1 only), likely due to non-overlapping regions (e.g., epitopes on trimer outside of S1); exposed regions that are available in the recombinant form but not in the complex (e.g., S1 in trimer complex versus S1 in recombinant form); and and conformational changes from the native or complex form versus the recombinant form.
The goal of the tutorial is to identify overlapping regions of interest (ROI), particularly at the heavy chain complementarity determining region (HCDR3), which is most often implicated in binding specificity of the antibody to the target. In particular, we will use the HCDR3 to identify the output populations that overlap across the RBD, S1, and trimer populations; we hypothesize the population at the intersection is likely to exhibit optimal binding profiles in the native conformation of the RBD in vivo, which will result in more optimal neutralization (RBD) potential relative to those HCDR3s that only recognize recombinant versions of the RBD or S1. Further, we want to ensure that antibodies display more favorable NGS metrics: high relative abundance, elevated fold enrichment, increased copy number of the full-length antibody, and reduced number of liabilities that may present problems during scaling up processes.
STEP 1 - Log in to Orion, Set Up Directory, Locate Tutorial Files
If the sequencing provider provides FASTQ files that are compressed (e.g., ending with “.gz” or “.zip”), decompress with a standard decompressor so the file ends with .fastq. If any suffix is added to the end, make sure to modify these suffixes so they end in .fastq for all sequence files to be processed.
Log in to the Orion interface with your email and password. Find the three required files under Organization Data as follows:
A) Organization Data / OpenEye Data / Tutorial Data / AbXtract / pacbio_small_codon_optimized.fastqB) Organization Data / OpenEye Data / Tutorial Data / AbXtract / barcode_file_abxtract.xlsxC) Organization Data / OpenEye Data / Tutorial Data / AbXtract / scaffold_ref_db_codon_optimized_dna.txtCreate a general tutorial directory and Tutorial 1 subdirectory under Project Directory / Tutorials / TUTORIAL_1 (This is your base directory and should be used for all outputs for this Tutorial 1 below).
STEP 2 - Select the ‘NGS Pipeline’ Floe
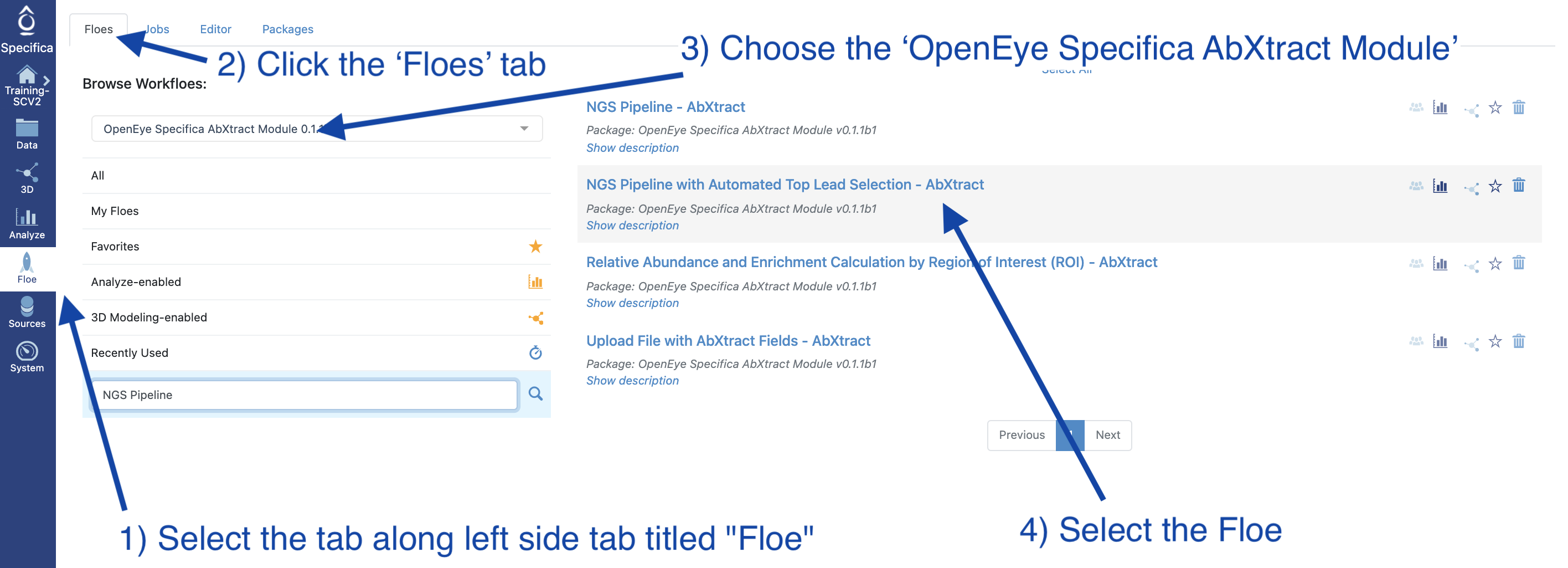
Figure 1. Select the appropriate floe.
Select the Floe page from the blue navigation bar.
Click the Floes Tab.
Under Packages, choose the OpenEye Specifica AbXtract Module.
Select the NGS Pipeline - AbXtract Floe.
STEP 3 - Prepare PacBio Run and Start Job
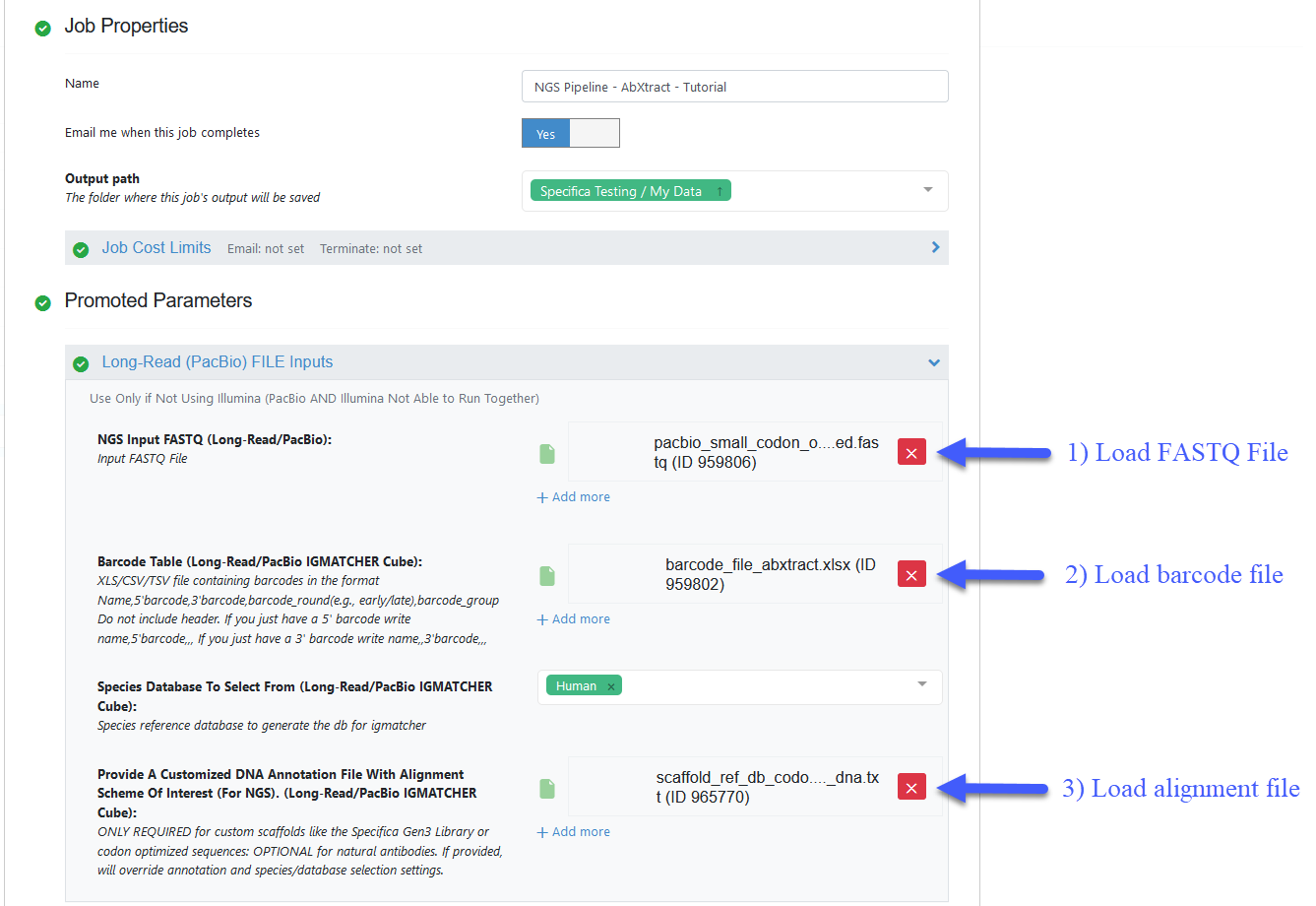
Figure 2. Job Form showing the PacBio inputs.
Load the FASTQ file - pacbio_small_codon_optimized.fastq.
Load the barcode XLSX file - Organization Data / OpenEye Data / Tutorial Data / AbXtract / barcode_file_abxtract.xlsx.
Load the alignment TXT file - Organization Data / OpenEye Data / Tutorial Data / AbXtract / scaffold_ref_db_codon_optimized_dna.txt.
Important Note
All remaining parameters can be kept as the default values. Scroll through remaining Promoted Parameters to get a sense of the options.
Click the “Start Job” button.
STEP 4 - Open the Floe Report to Get a General Idea About the Population
On the Floe page and under the Jobs Tab, select the NGS Pipeline Floe.
In the Job Panel, you can find job details, a results drop-down (connecting to datasets and collections), and a report drop-down (Floe Report). These are shown in Figure 3.
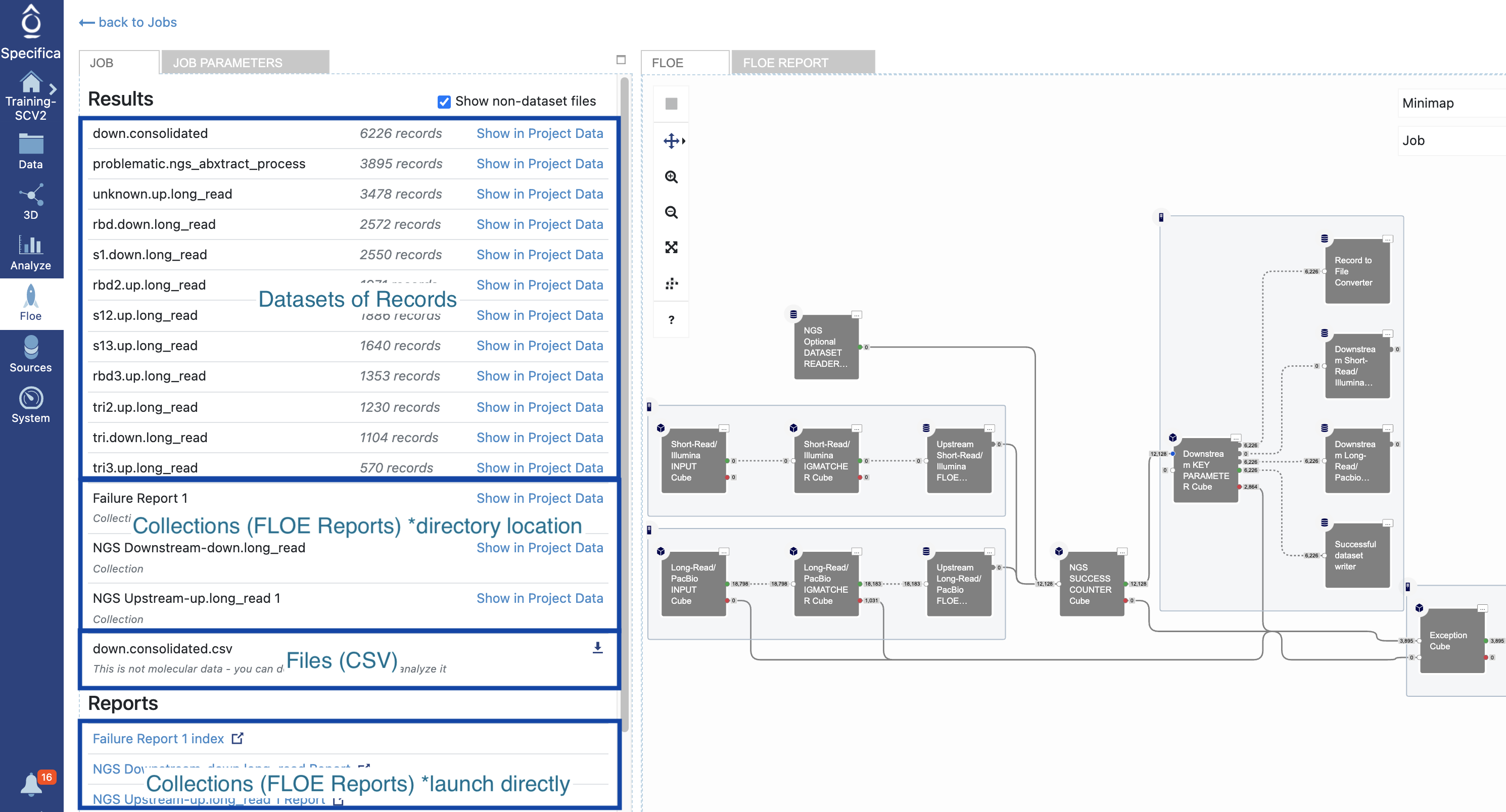
Figure 3. Job Panel.
Click on the NGS Downstream-down.long_read file under the Reports section to see the Floe Report. Alternatively, you can click on the Floe Panel to the right.
The General Stats table provides a snapshot of the number of nonredundant full-length LCDR3, HCDR3, LCDR3 and HCDR3 sequences. Chain_1 represents the variable light chain (VL) and chain_2 represents the variable heavy chain (VH). The overlap shows the overlap of the region of interest (HCDR3) across the different populations.
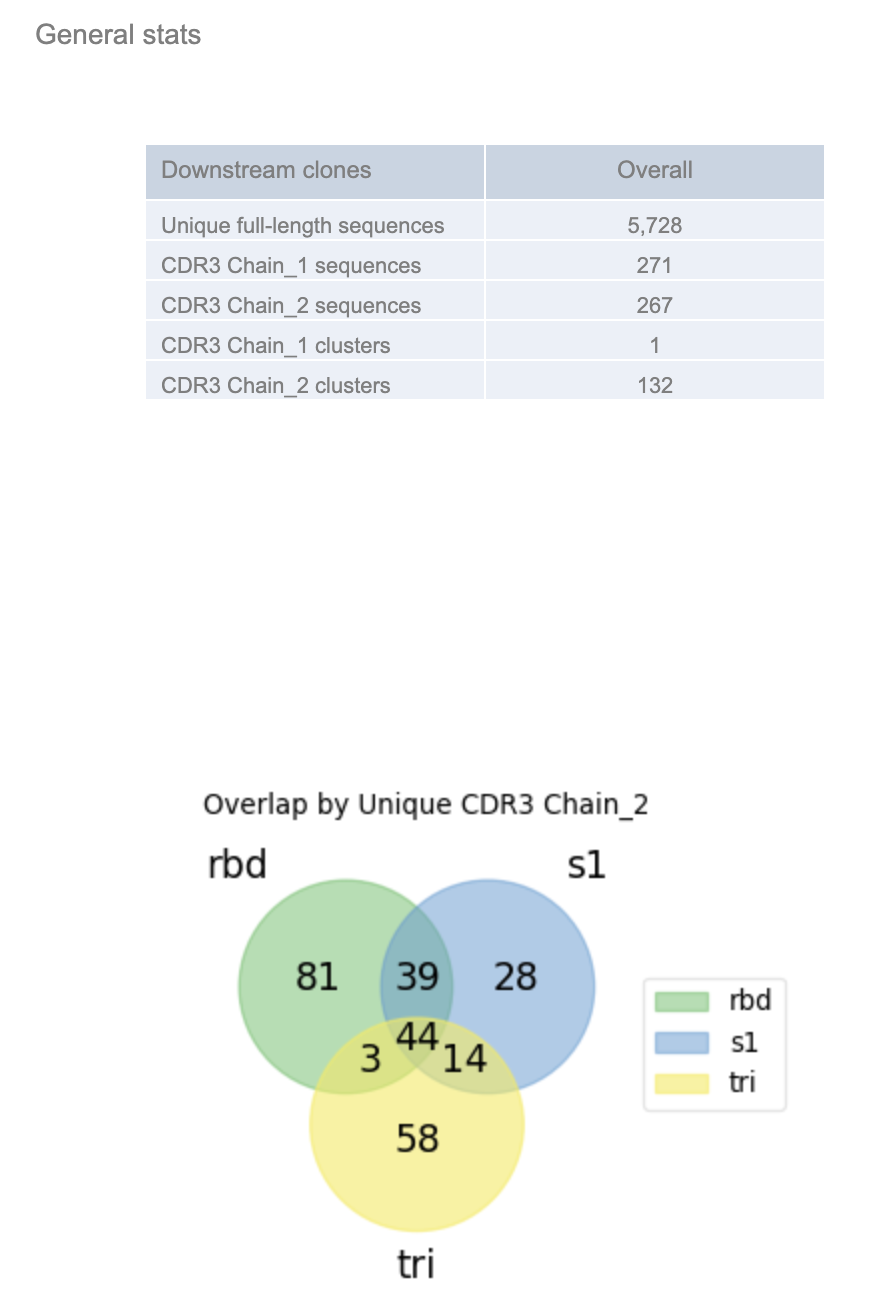
Figure 4. General Stats in Floe Report.
Each subpanel separates out the barcode_group and shows the population-specific (e.g., trimer, S1, RBD) statistics.
Return to Job Panel (see see Figure 3.)
Under the Results drop-down, find the dataset called down.consolidated and click “Show in Project Data” to go directly to the Data page.
STEP 5 - Activate the Dataset
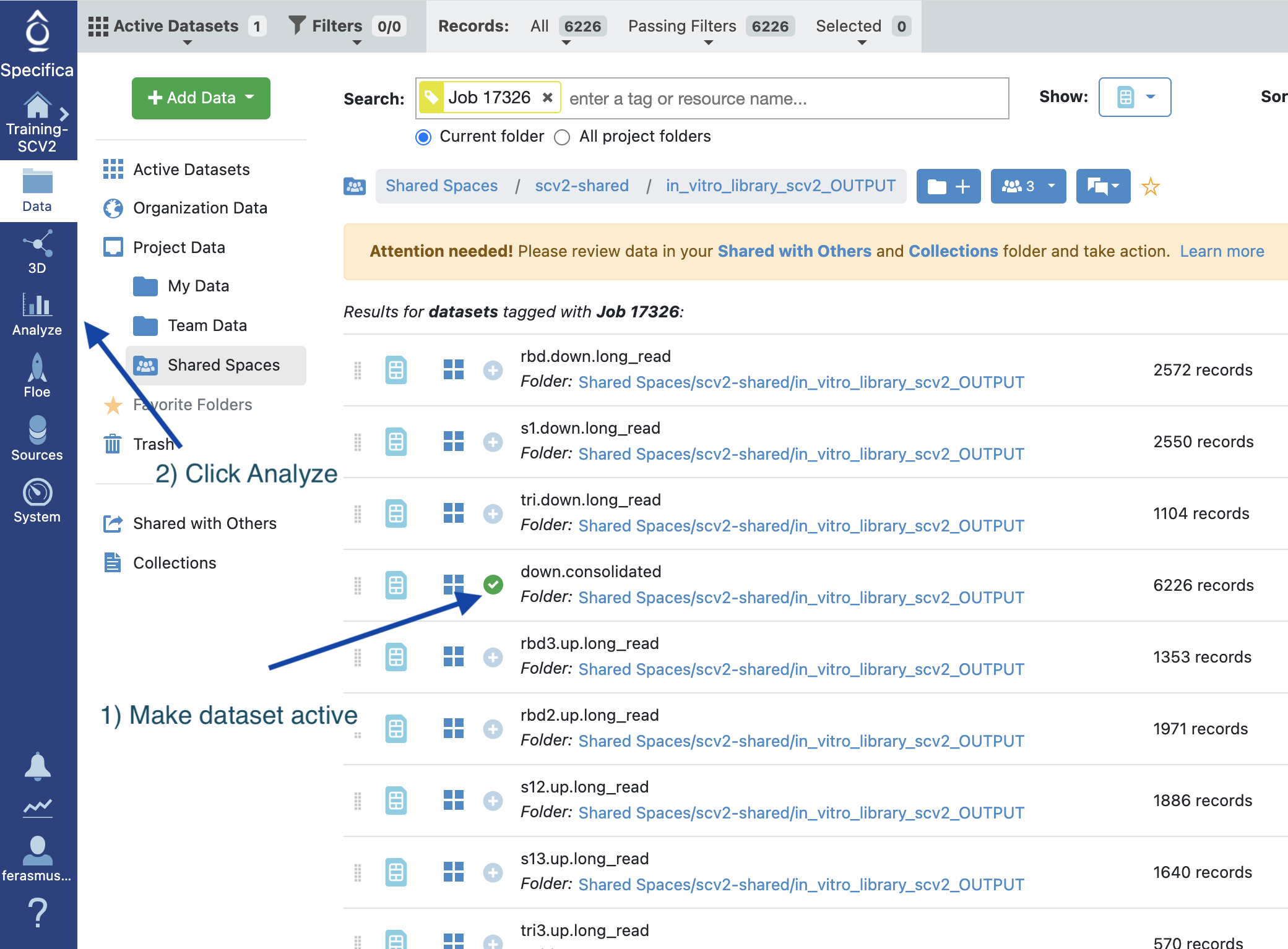
Figure 5. Activate the Dataset.
Select the “+” symbol next to the down.consolidated to activate the dataset.
Select the Analyze page.
STEP 6 - Plot Relative Abundance Versus Desired Overlap Population and Select Population with Analyze Tool
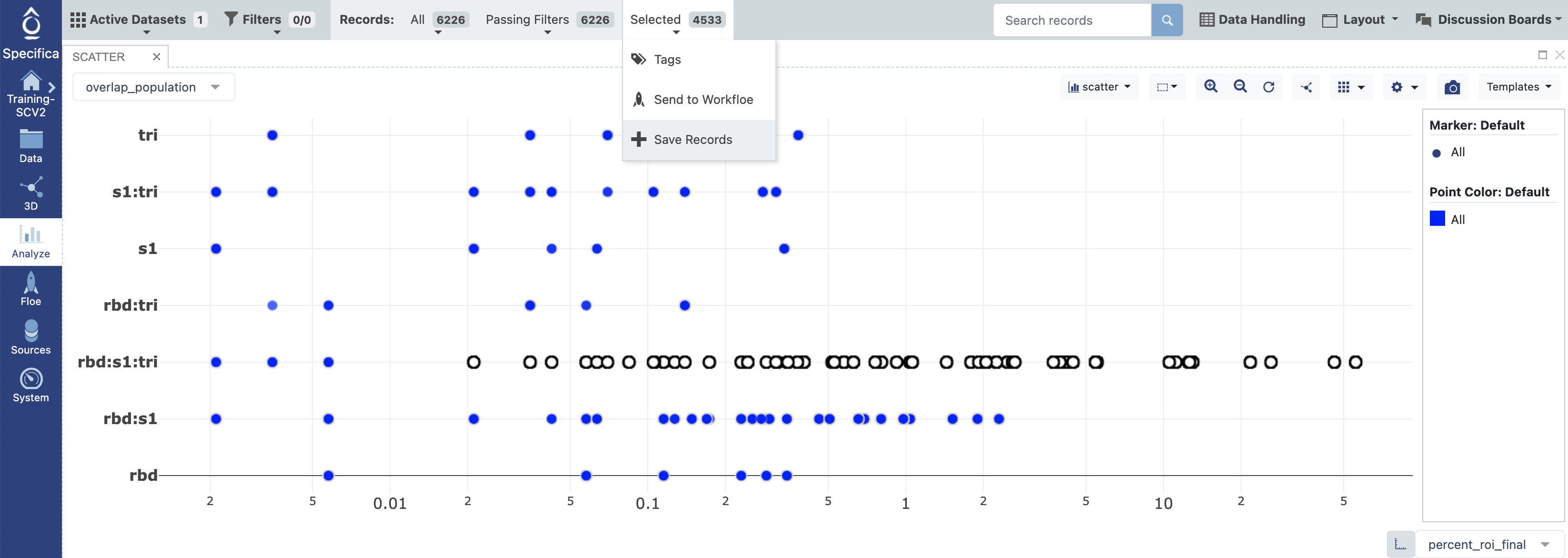
Figure 6. Analyze page.
Select percent_roi_final for the x-axis.
Select overlap_population for the y-axis.
Subselect records within the rbd:s1:trimer population with an ROI above the 0.01% as shown above.
On the Active Data Bar at the top of the page, the ‘Selected’ drop-down will show the number of selected antibodies. Click on this and then choose the “Save Records” button.
In the box that appears, choose “New Dataset,” name it rbd_s1_trimer>0.01, select the “Do Nothing” option, and choose your Base Directory (see see STEP 1, #3.).
Allow some time to generate the new dataset of ~4.5K records. Find the new dataset in the specified directory. If it needs additional time to process, the dataset may be grayed out. If it remains grayed out, select the “…” ellipses to the right of the dataset and choose the “Process Data” option.
Remove any active datasets by selecting selecting “Clear All” in the ‘Active Datasets’ drop-down.
STEP 7 - Select the Enriched Population Using the Analyze Tool and Send to the Automated Top Lead Selection Floe
Make the newly created rbd_s1_trimer>0.01 dataset active.
Choose the “Data Handling” option on the Active Data Bar.
Make sure that the log2_enrichmment_roi is active across all options. Click the “Apply” button.
On the x-axis, select log2_enrichment_roi. If it doesn’t appear in the drop-down, use the default axes, then immediately cancel. The log2_enrichment dataset should now appear in the drop-down menu for x-axis parameters. For the y-axis, choose any parameter (e.g., overlap_population).
Choose records with log2_enrichment of ≥ 1. Find the field log2_enrichment_roi in the table, select the down arrow, and click ‘sort descending.’
Select all rows having a log2_enrichment value of ≥ 1; the selected population should be selected in the plot in Figure 7.
Click the ‘Selected’ drop-down on the Active Data Bar and choose Send to Workfloe as shown in the image.
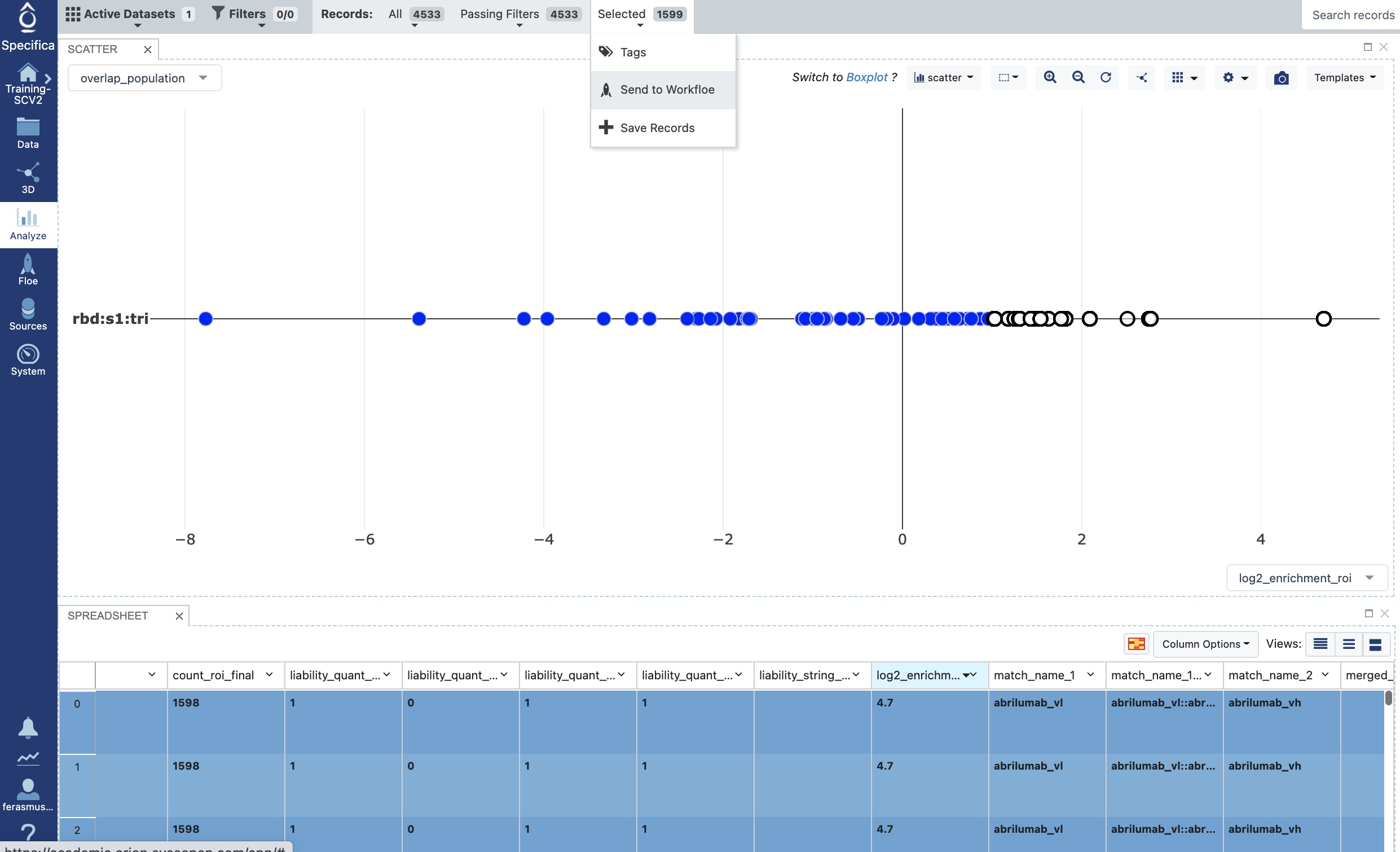
Figure 7. Select the enrichment population.
On the Floe page, search for the Automated Top Lead Selection Floe and click the “Launch Floe” button.
Set the Output path to the Base Directory (see STEP 1, #3).
Use default values for the NGS Key Selection Parameters.
Description of Key Selection Parameters
Maximum Number Of Full-Length Sequences: The maximum number of full-length, nonredundant sequences to output.Maximum Number Sequences per Cluster: The maximum number of unique full-length sequences per given cluster.Maximum Number Of Clusters Preferred: The maximum number of clusters that we want to select from.Metrics For Ranking: Metrics that determine how the sequences will be sorted in output.Rank Sanger Clones First In Population: Keeps SANGER population (ranked like Metrics for Ranking) at the top of the rank order. Only used if the Sanger dataset/file is provided.Attempt To Fill The Desired Number Of Full-Length Sequences Quota: Attempts to fulfill the total number of sequences (from Maximum Number Of Full-Length Sequences) by selecting additional full-length sequences from the same clusters, followed by selecting the remaining top-ranked clones from different clusters.
Keep the default output names.
Start the job.
STEP 8 - Subset the Number of Fields for Output and Download CSV
Find the picked.consolidated dataset created in STEP 7 either on the Data page or directly from the Results drop-down on the Job Panel (click “Show in Project Data”).
On the Data page, click the “Send to Workfloe” icon for the picked.consolidated dataset. Next, on the Floe page, find the Subset the Number of Fields for Export Floe and click the “Launch Floe” button.
Keep these fields for the following parameters (for more details, see key fields reference):
A) Identifier Fields to Keep: seq_idB) Sequence Fields to Keep: sequence_aa_1, sequence_aa_2, match_name_1, match_name_2C) Cluster Fields to Keep: clusterUse the following names for the output datasets:
A) Output Dataset: gene_synthesis_clones_with_cluster_idB) Output CSV Filename: gene_synthesis_clones_with_cluster_id.csvClick the “Start Job” button.
After the floe is finished, in the Jobs Tab, click on the job for the Subset the Number of Fields for Export Floe. Open the job to view datasets, as follows:
Under the Results drop-down on the Job Panel, find the CSV file gene_synthesis_clones_with_cluster_id.csv.
Download the file to obtain the CSV list of sequences ready for gene synthesis.