Antibody SiteHopper-based Clustering Tutorial
SiteHopper compares binding sites on the protein surface by looking at their shape and chemical features. For antibodies, we focus on the shape and chemical features of the region around the CDR regions. A much higher emphasis is placed on the chemical features (75%) than the shape (25%) for the calculation of the SiteHopper score. The SiteHopper results for these regions are compared for a dataset of 3D antibodies to identify clusters, which are expected to have similar binding properties.
The Antibody SiteHopper-based Clustering Floe requires a dataset of 3D antibody structures. This dataset can be generated using the Antibody Sequences to 3D Models Floe and the Antibody Experimental Structure Prep Floe.
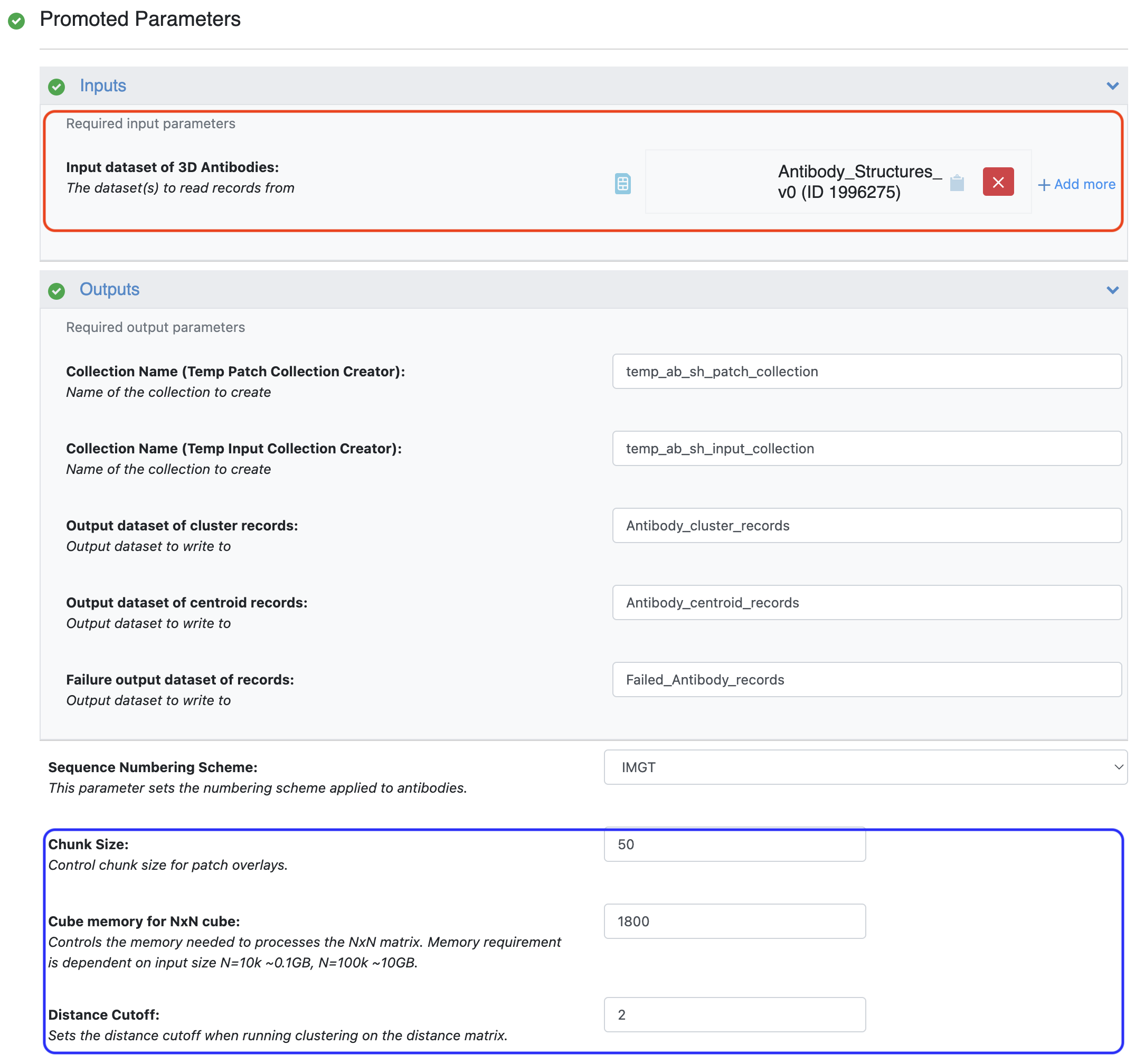
Figure 1. Inputs and parameters for the Antibody Sitehopper-based Clustering Floe. The input region is annotated with a red box and the parameter region is annotated with a blue box.
The floe generates a score for each pairwise comparison of the antibodies in the dataset. The Cube Memory for NxN Cube parameter must be specified based on the size of the dataset under consideration. A lower than required number will cause the floe to fail. The Distance Cutoff parameter specifies the SiteHopper score threshold below which binding sites are considered similar for the clustering algorithm. The SiteHopper score has a maximum value of 4.0.
Result Analysis
The floe generates a cluster records dataset which contains the cluster identity of each antibody in the dataset, and a centroids dataset which only contains information on the centroids. These can be visualized in the 3D Viewer if needed. As shown in Figures 2 and 3, the surface patch can be observed to compare antibodies. Antbodies belonging to different clusters are shown in Figure 2 where the surface of the CDR2 region can be observed to be very different between the two antibodies. In Figure 3, on the other hand, the antibodies belong to the same cluster and the surface of the CDR2 region is very similar.
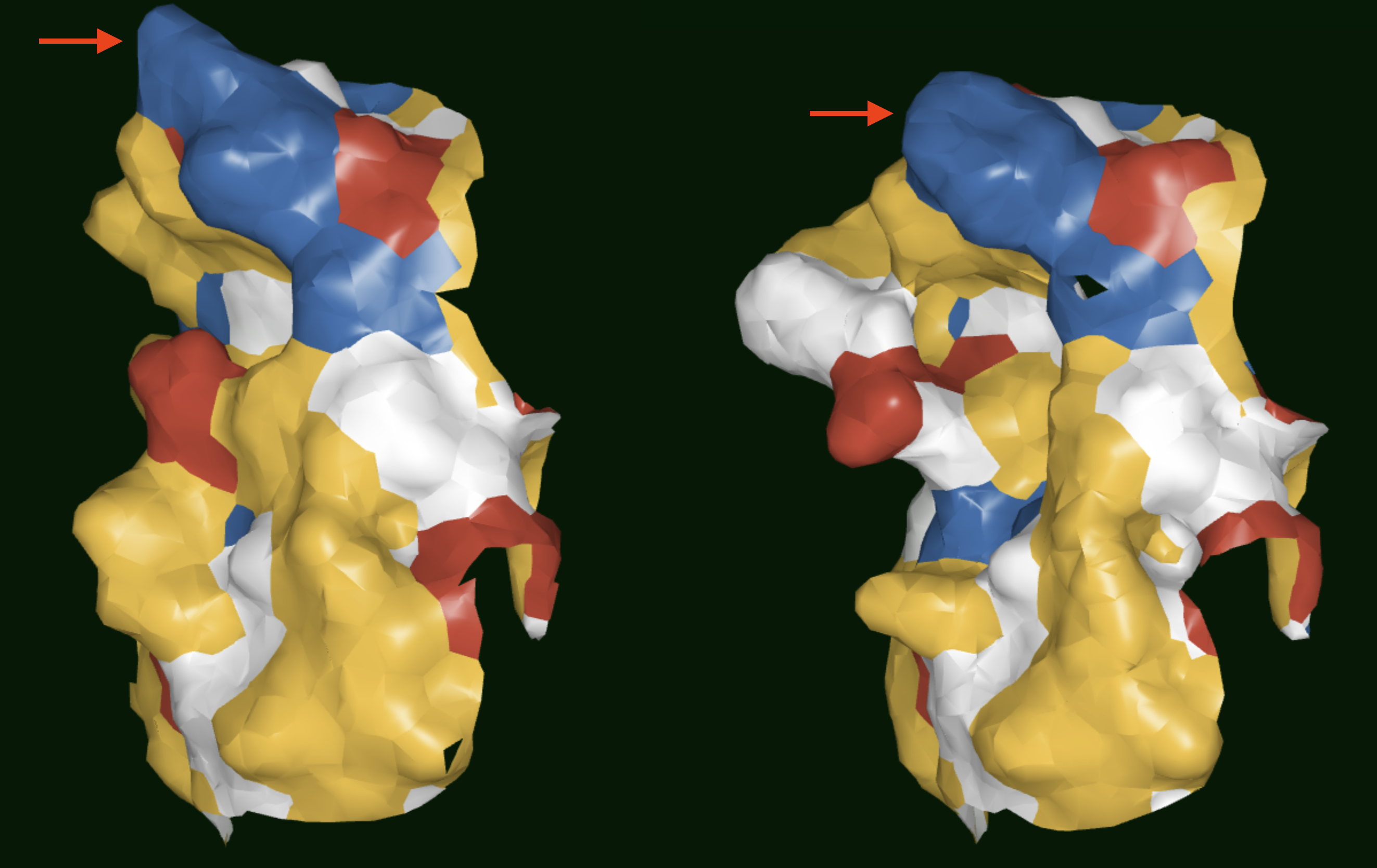
Figure 2. Results showing the surface patches of antibodies belonging to different SiteHopper-based clusters. Red arrows point to the CDR2 regions.
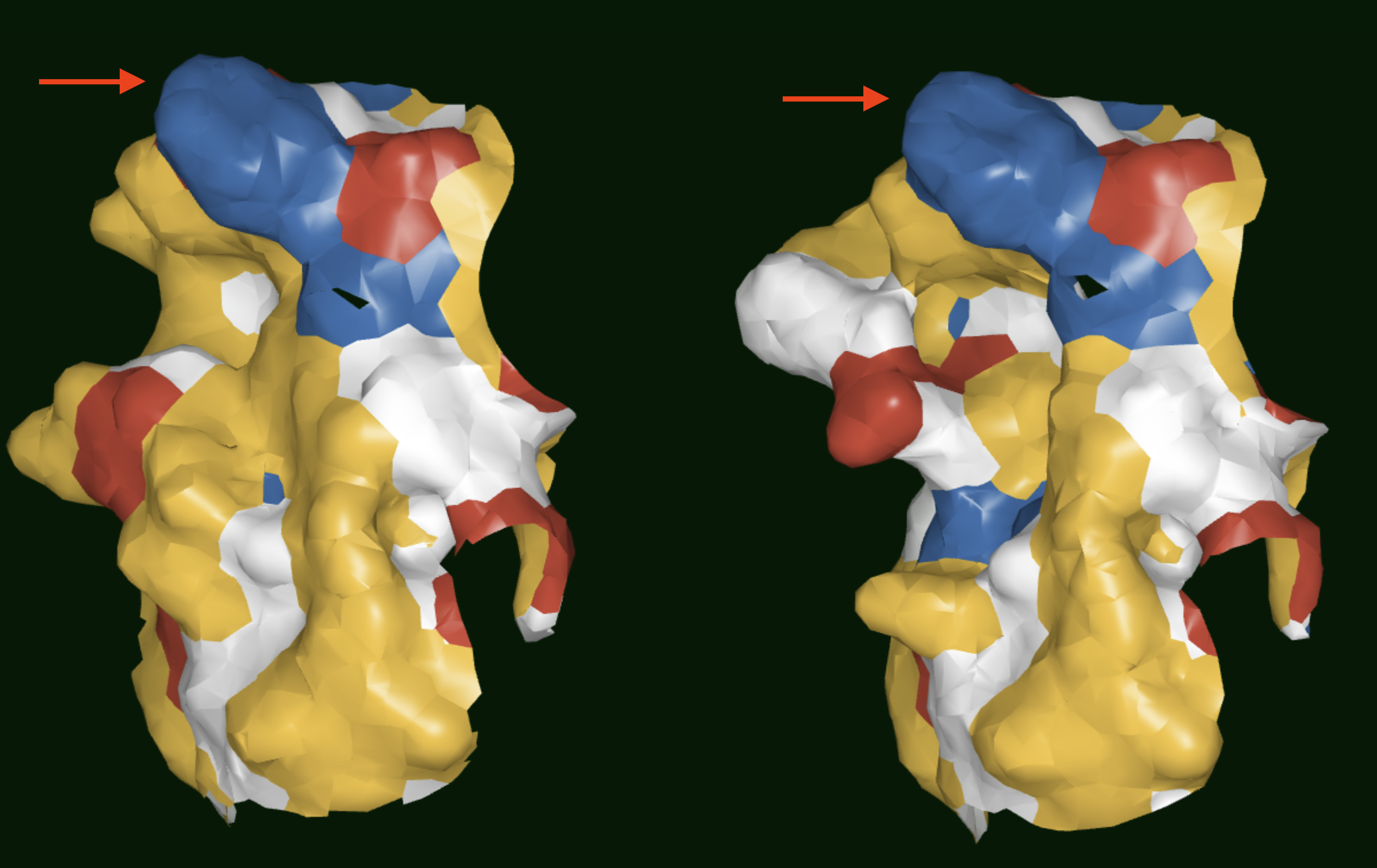
Figure 3. Results showing the surface patches of antibodies belonging to the same SiteHopper-based cluster. Red arrows point to the CDR2 regions.