Understanding Antibody Surfaces and Annotations
Surface Patches
Different floes add surface hydrophobicity and charge patches to the antibody models that can then be viewed in the 3D Viewer. In the case where the structures do not have the mentioned patches, they can be added using the Antibody Surface Patch Generation Floe.
Hydrophobicity of the surface can be visualized by toggling the hydrophobic surface options in the drop-down menu for an item in the Data section of the 3D Viewer (Figure 1). Jain SAP scores are used by default. Hydrophobic regions show up as green in the Viewer, while hydrophilic regions show up as purple. Side chains are colored green if they participate in a hydrophobic patch.
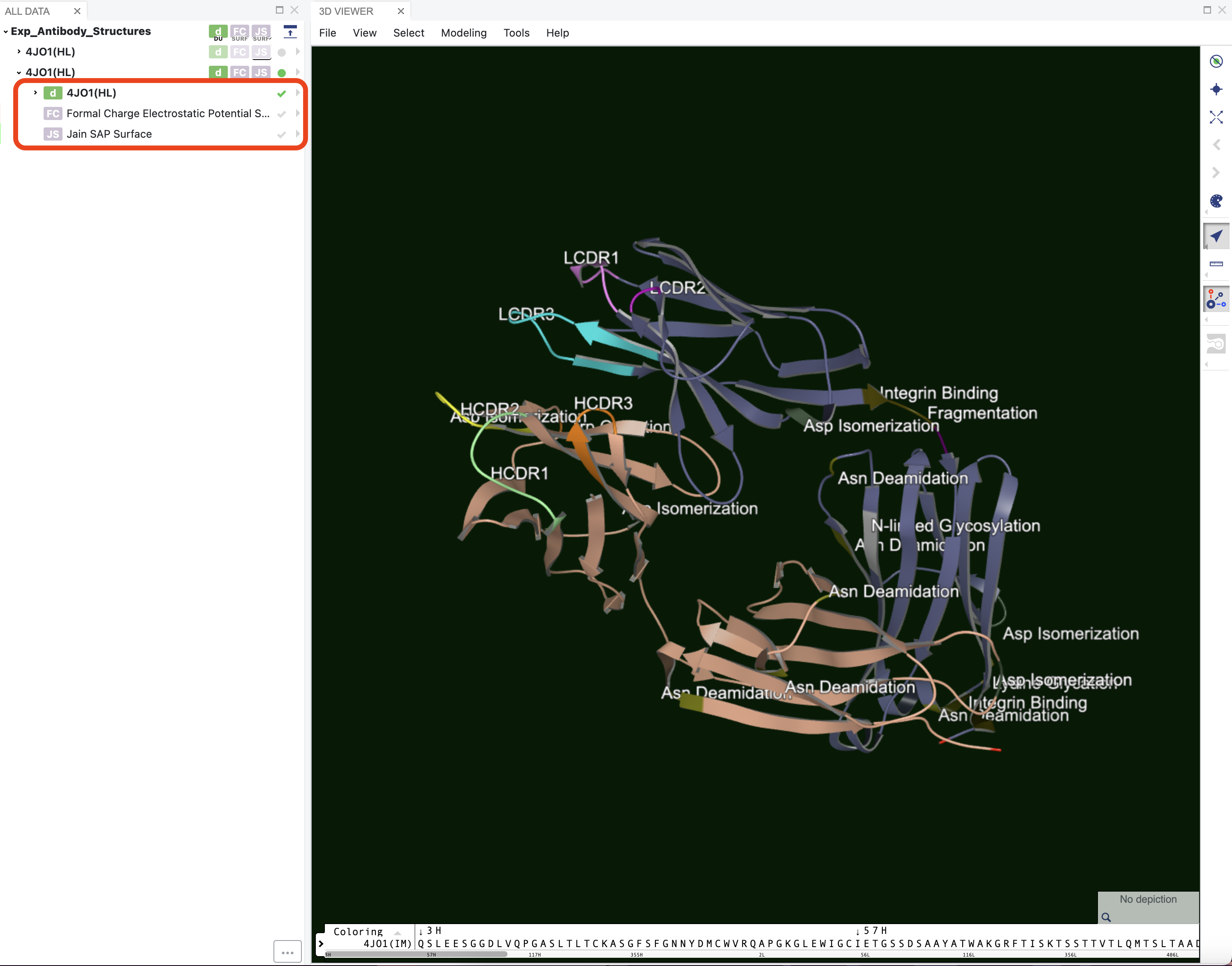
Figure 1. Drop-down menu for an item in the 3D Viewer showing the “Formal Charge Electrostatic Potential” and “Jain SAP Surface” surface visualization options.
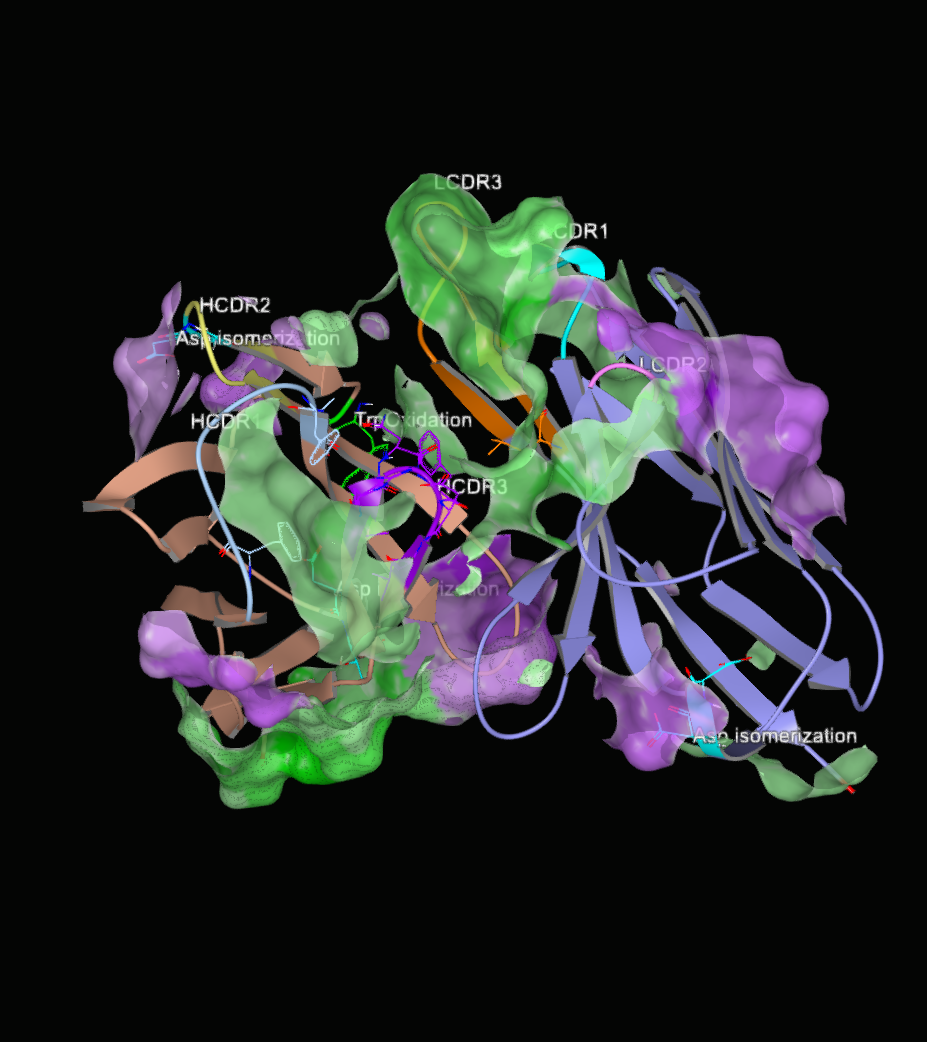
Figure 2. Hydrophobicity patches on the surface. Opacity of the surface was set to 75%.
The following references offer hydrophobicity scoring schemes that are available for analysis.
Surface charge can also be visualized on the antibody structure. It can be toggled using the “Formal Charge Electrostatic Potential Surface” option in the drop-down menu for an item in the 3D Viewer. Negatively charged areas show up as red, while positively charged areas show up as blue.
Figure 3. Charge patches on the surface. Opacity of the surface was set to 75%.
Patches and styling can be changed using the Antibody Surface Patch Generation Floe if needed.
Structure Annotations
Along with surface patches, the antibody modeling floes also add structural annotations that mark the CDR regions and areas where liabilities are at the antibody surface including motifs associated with post and co-translational modifications. The CDR regions are determined by numbering the sequence using ANARCI [Dunbar-2015], while the liabilities are found by searching for associated sequence motifs and verifying that the relevant residues in those sequence motifs are exposed to the surface of the antibody protein. The locations of the following liabilities can be annotated where they are applicable.
Lysine Glycation
CD11e/CD18 Binding
Asn Deamidation
Fragmentation
Met Oxidation
Asp Isomerization
Integrin Binding
Trp Oxidation
Unpaired Cyst
N-linked Glycosylation
N-terminal Glutamates
Figure 4 shows the model for 4JO1 built using its sequence. The annotations include the CDR regions for both the heavy and the light chains. It also shows areas of predicted chemical liabilities.
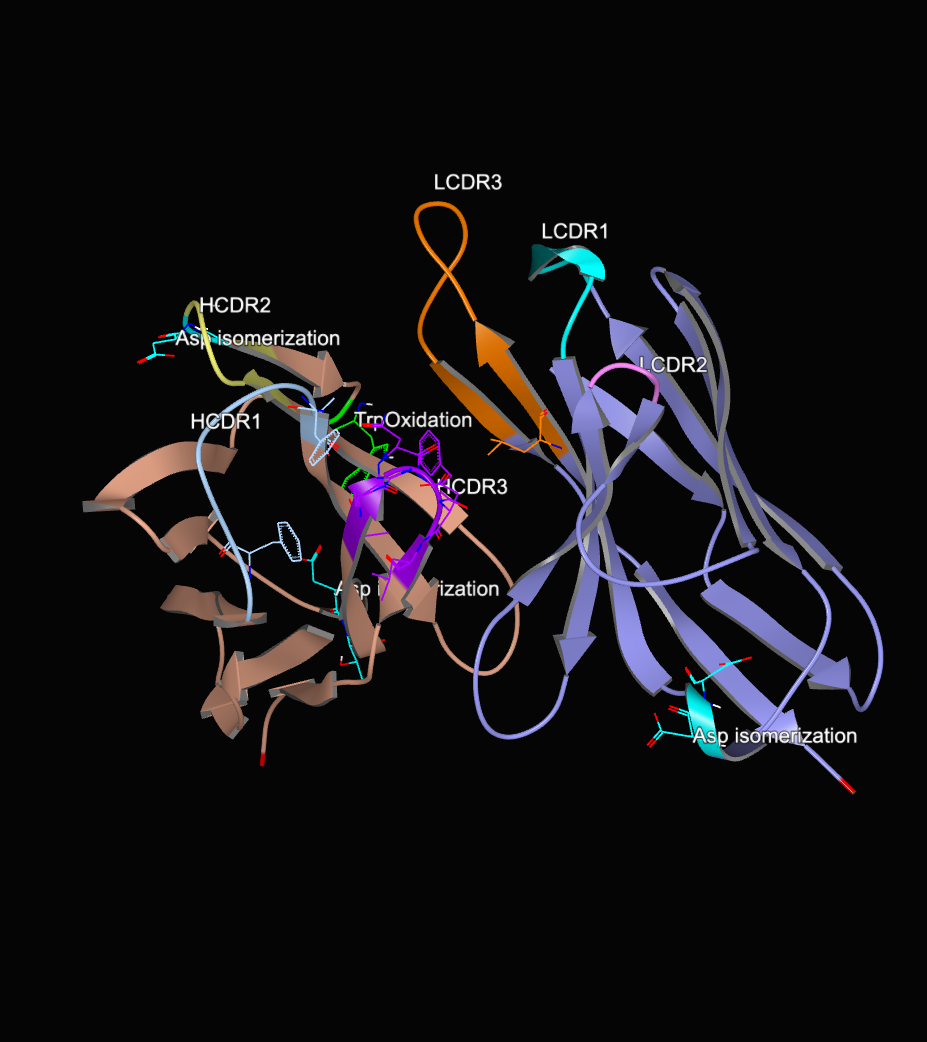
Figure 4. Annotated structure of an antibody.