Release Highlights 2025.4
Small Molecule Discovery Suite
Small Molecule Modeling Floes
Figure 1. New clustering floes support large-scale datasets.
This release introduces three new clustering floes for large-scale compound clustering. These floes use different clustering strategies based on the type and size of data to be clustered. The Clustering: 2D Scaffold (~25M compounds) Floe is well suited for high-throughput virtual screening. It uses Bemis-Murcko scaffolds and can be used to cluster up to 25 million compounds. The Clustering: 2D Fingerprint, up to 100K compounds Floe uses a variety of fingerprint types to subsample diverse datasets of up to 100,000 molecules. For 3D datasets, the Clustering: 3D Shape/Color, ~10K compounds Floe uses shape and color similarity to cluster datasets of tens of thousands of molecules. These floes have been optimized for performance and scalability on Orion with significantly reduced run times compared to previous implementations. Additionally, the 2D Fingerprint and 3D Shape/Color Floes generate intermediate collections of pairwise distances, allowing iterative clustering runs with varying parameters to be completed in a timely and cost-efficient manner. These floes can support virtual screening at the largest scale and guarantee selection of a diverse list of compounds for downstream applications.
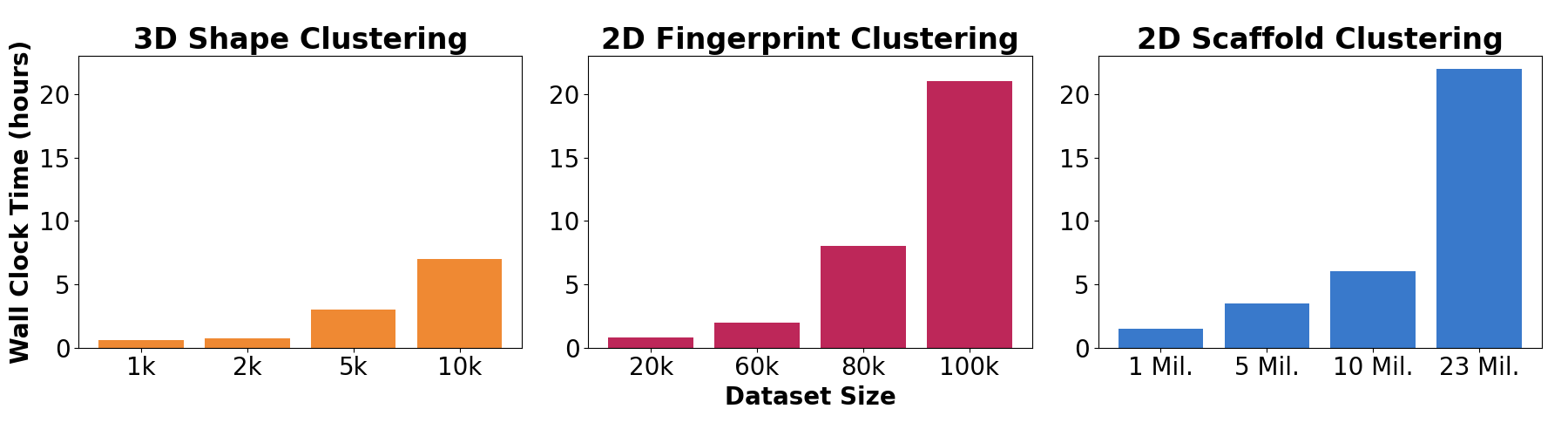
Figure 2. Wall clock times (y-axis) for various dataset sizes (x-axis) for each of the three new clustering schemes, increasing from left to right in dataset size: 3D Shape (orange), 2D Fingerprint (red), and 2D scaffold (blue).
AI Fold Floes
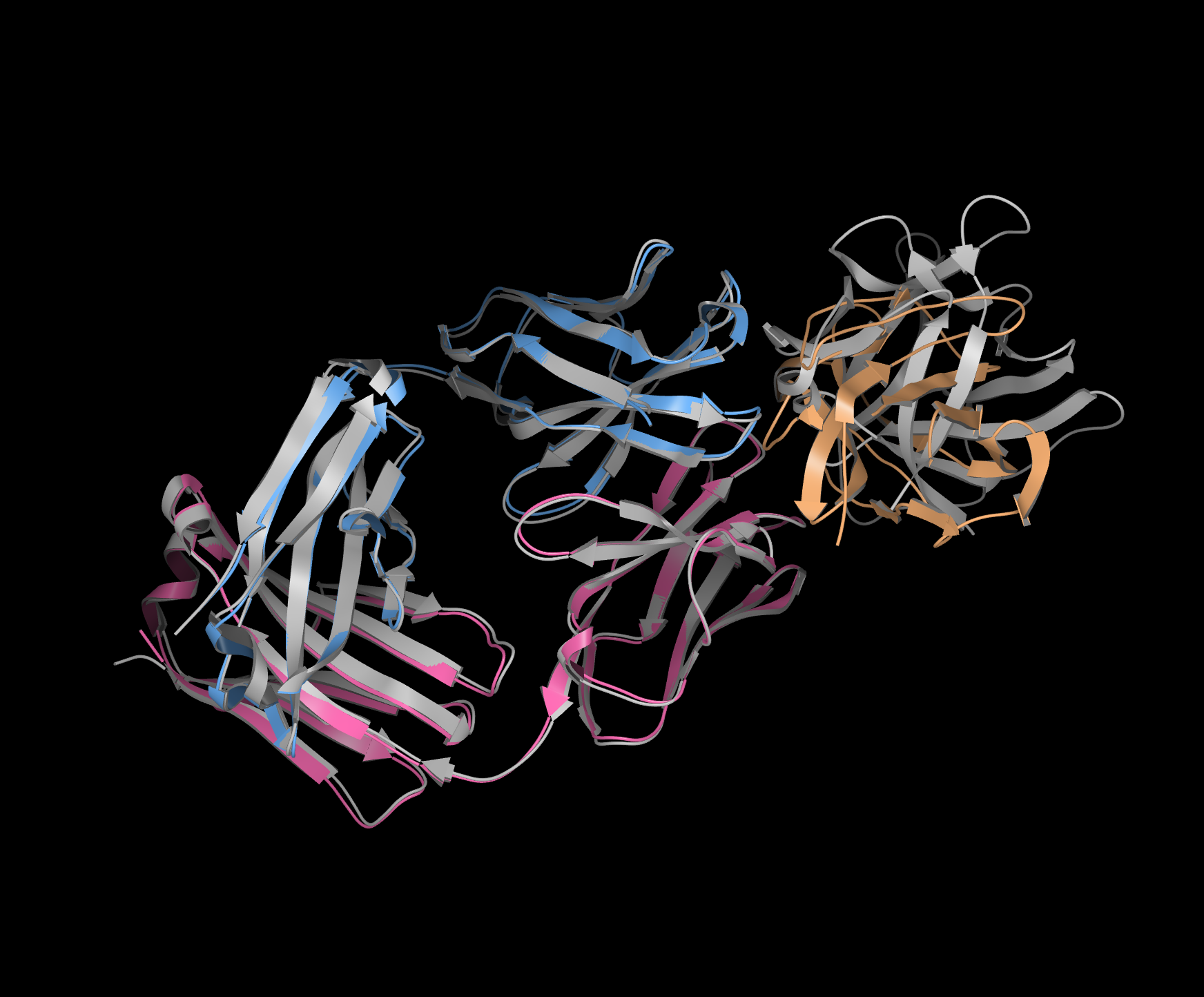
Figure 3. Boltz-2 used to predict human IL-1beta bound to the antibody binding fragment of canakinumab, showing the experimental structure (PDB 4G6J) in grey and the predicted structure with chain A (tan), chain B (blue), and chain C (pink).
The AI Fold Floes have been upgraded to replace OmegaFold with Boltz-2. The multi-sequence alignment portion of the floes has been optimized for Orion architecture to reduce computational cost and time. The Protein Sequence to AI Folded Structure Prediction Floe now supports co-folding protein–ligand complexes and predicts structures of multimeric protein complexes. The new Protein Sequence to AI Folded Structure Ligand Affinities Floe predicts the co-folding structure of a provided list of ligands for the same protein, resulting in a hit list of complexes based on predicted affinity. Multiple constraint schemes, including bonding patterns for covalent ligands and pocket definitions for co-folding, use knowledge of the system to get the best predicted structures possible. A tutorial has been added to accompany the new floes.