Release Notes, Version 2025.1.2
Highlights of 2025.1.0
Floe Release 2025.1.2 is a bug fix release with improvements made to Cryptic Pocket Detection, Large Scale Floes, Molecular Dynamics Affinity Floes, and Molecular Dynamics Core Package.
Molecular Dynamics Affinity Package
Improved Interactive Edge Mapper: The Edge Mapper for RBFE Calculations Floe generates pragmatic maps of the relative affinities to be computed by NES. An interactive floe report is now available, allowing you to adapt this map to your needs and save the changes on Orion. In this report, you can easily add a new edge, delete an edge, or make a new star map with the selected ligand as a hub. This tutorial shows how to use the floe and manipulate the data from the Floe Report. Figure 1 shows the Floe Report edge map, as well as some ways to edit it. The new edge map can be saved as a dataset for downstream free energy calculations.
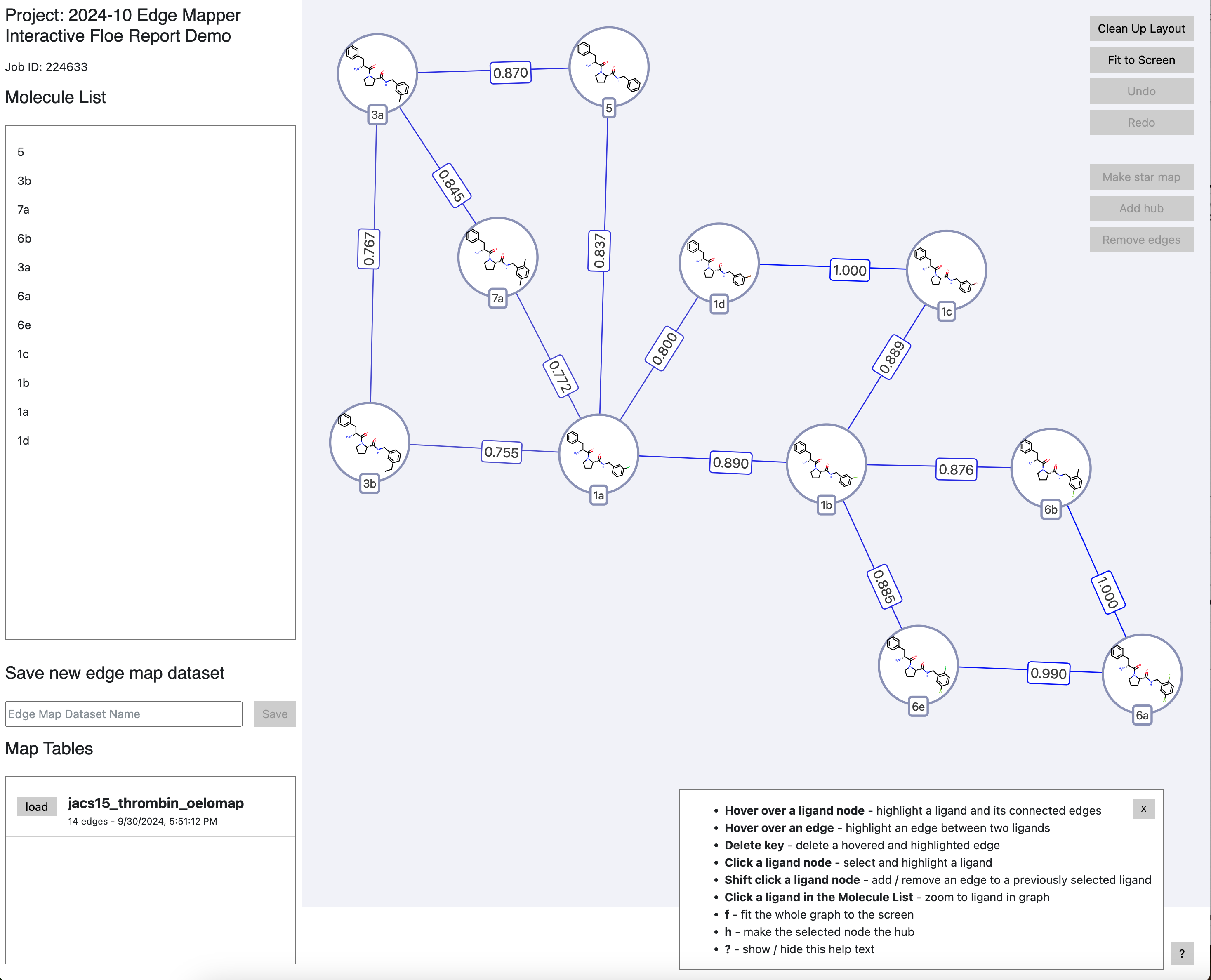
Figure 1. The interactive edge map Floe Report for the Edge Mapper for RBFE Calculations Floe.
New Map Option: A new clustering-based edge map (multi-star map) has been added. The hub selection scheme for the star map has been updated for improved robustness.
3D QSAR Modeling Floes
Updated 3D QSAR Consensus Model: The consensus model for the 3D QSAR Floe package has been updated to a weighted model that averages all included models, including 2D as a baseline. The new weighted model consistently outperforms the previous unweighted model, as can be seen in Figure 2.
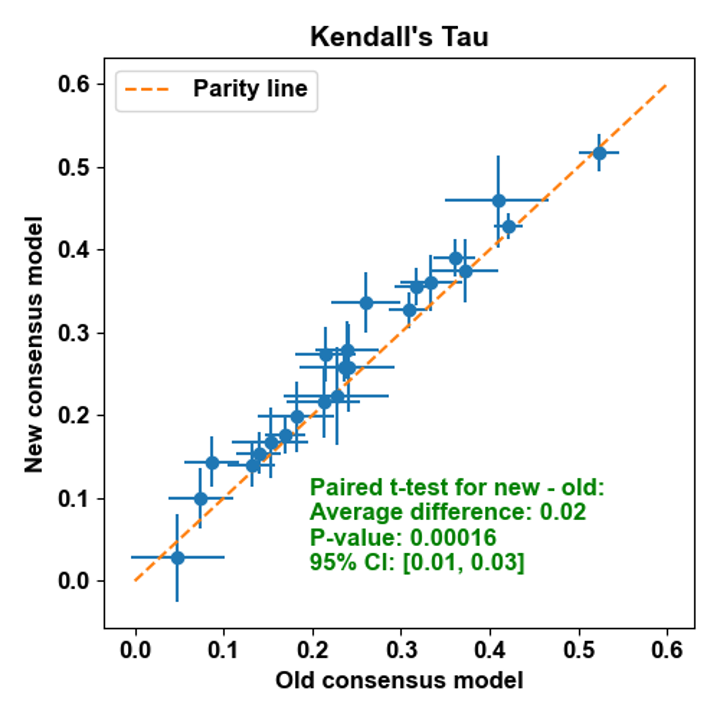
Figure 2. A comparison of Kendall’s Tau using the new consensus model versus the old model.
Improved 3D QSAR Method: The 3D QSAR approach for deriving a molecule now substantively outperforms most potent molecule queries, as can be seen in Figure 3.
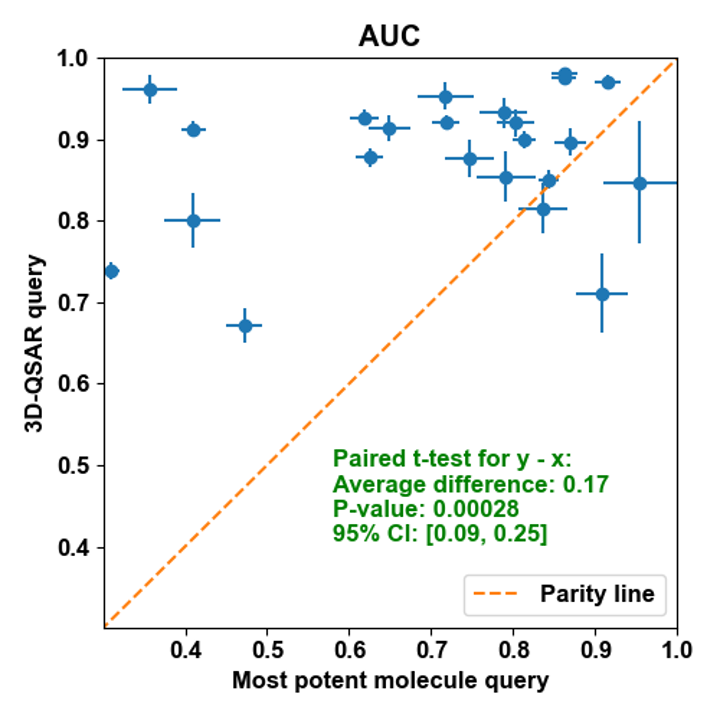
Figure 3. A comparison of AUC of the 3D QSAR-ROCS query versus the most potent molecule queries from the training set.
Orion Floes
The Orion floes have been reorganized to mirror the drug discovery process. The Small Molecule Discovery Suite includes three modules: Target Exploration, Hit Identification, and Lead Optimization. Most previous OpenEye floe packages exist within these new modules, although a few have been refactored into different packages or categories. The primary floe packages that were refactored include Biomodeling Floes, Cheminformatics Floes, Classic Floes, and Large Scale Floes. Some floes now function across all modules.
Detailed Release Notes, Version 2025.1.2
The packages below have been updated for this release. The other packages are the same as shipped with release 2024.2.2.
- Legacy Release Notes
- ChemInfo Hit ID Release Notes
- Classic Lead Optimization Release Notes
- Legacy Release Notes
- Legacy Release Notes
- Release Notes
- Legacy Release Notes
- Large-Scale Floes Hit-to-Lead Release Notes
- Legacy Release Notes
- Legacy Release Notes
- Legacy Release Notes
- Protein Modeling Release Notes
- Small Molecule Modeling Release Notes
- Legacy Release Notes
- Legacy Release Notes