Spruce Prep Tutorial
The Spruce Protein Preparation Floes use Spruce to prepare biomolecular systems for use in downstream modeling applications. The floe generates one or more prepared design units with associated depictions, to illustrate the protein-ligand binding site interactions, as well as the Iridium classification.
The preparation steps of the floe include:
Expansion of the asymmetric unit to the biological unit, if necessary (e.g., if the structure is from an X-ray crystallography experiment). The biological unit is what is known or believed to be the form the protein(s) take in-vivo or in-vitro.
Enumeration (default) or collapse of alternate locations.
Building missing pieces, including partial side chains, modeling missing loops and tails, and capping chain breaks. To learn more about why Spruce may output partial residues, please see the Spruce TK theory section.
Placement and optimization of hydrogen atoms, including tautomer enumeration of ligands and cofactors, as well as evaluation of those tautomer states in the biomolecular structure.
The protein and ligand are assigned partial charges (AmberFF99SB and AM1BCC, respectively) and the output dataset is ready for downstream structure-based modeling tasks.
Floes Used in the Tutorials
The floe used in this tutorial is documented here:
There are two primary methods for protein structure input, and both are shown below. To run the SPRUCE - Protein Preparation Floe, navigate to the Floe tab in Orion and then:
Type SPRUCE in the search box.
Select the SPRUCE - Protein Preparation Floe and click the “Launch Floe” button to open the Job Form.
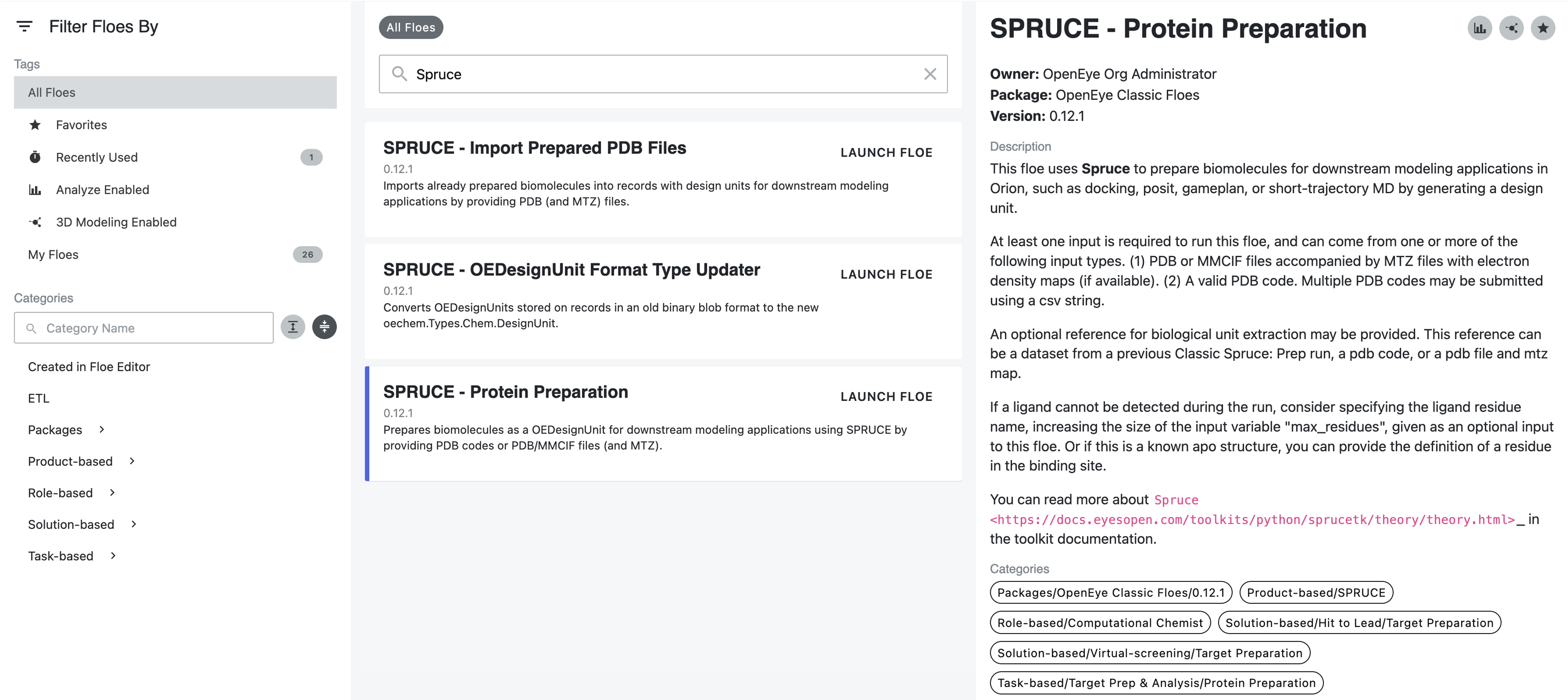
Figure 1. The Spruce Protein Preparation Floes.
The first method for protein structure input takes a list of PDB codes and pulls the structures and their associated electron density maps (MTZ files) from the PDB server hosted at the RCSB.
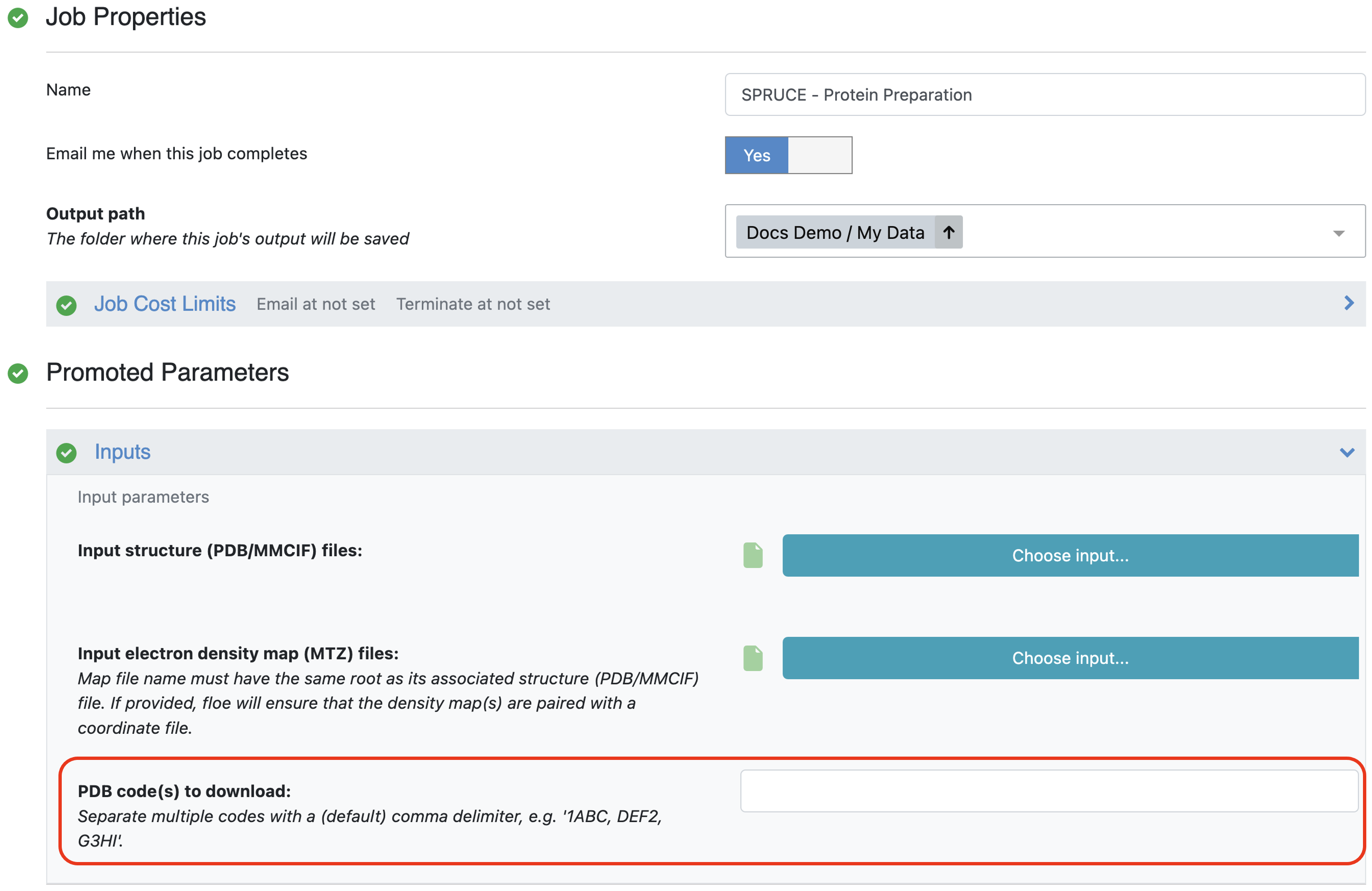
Figure 2. The area on the Job Form to provide PDB codes as input.
The second method accepts a list of PDB files. These can already be uploaded to Orion or can be uploaded as part of the job submission. We recommend providing the associated MTZ files to enable calculation of the Iridium classification and to provide electron density grids for structure inspection after preparation.
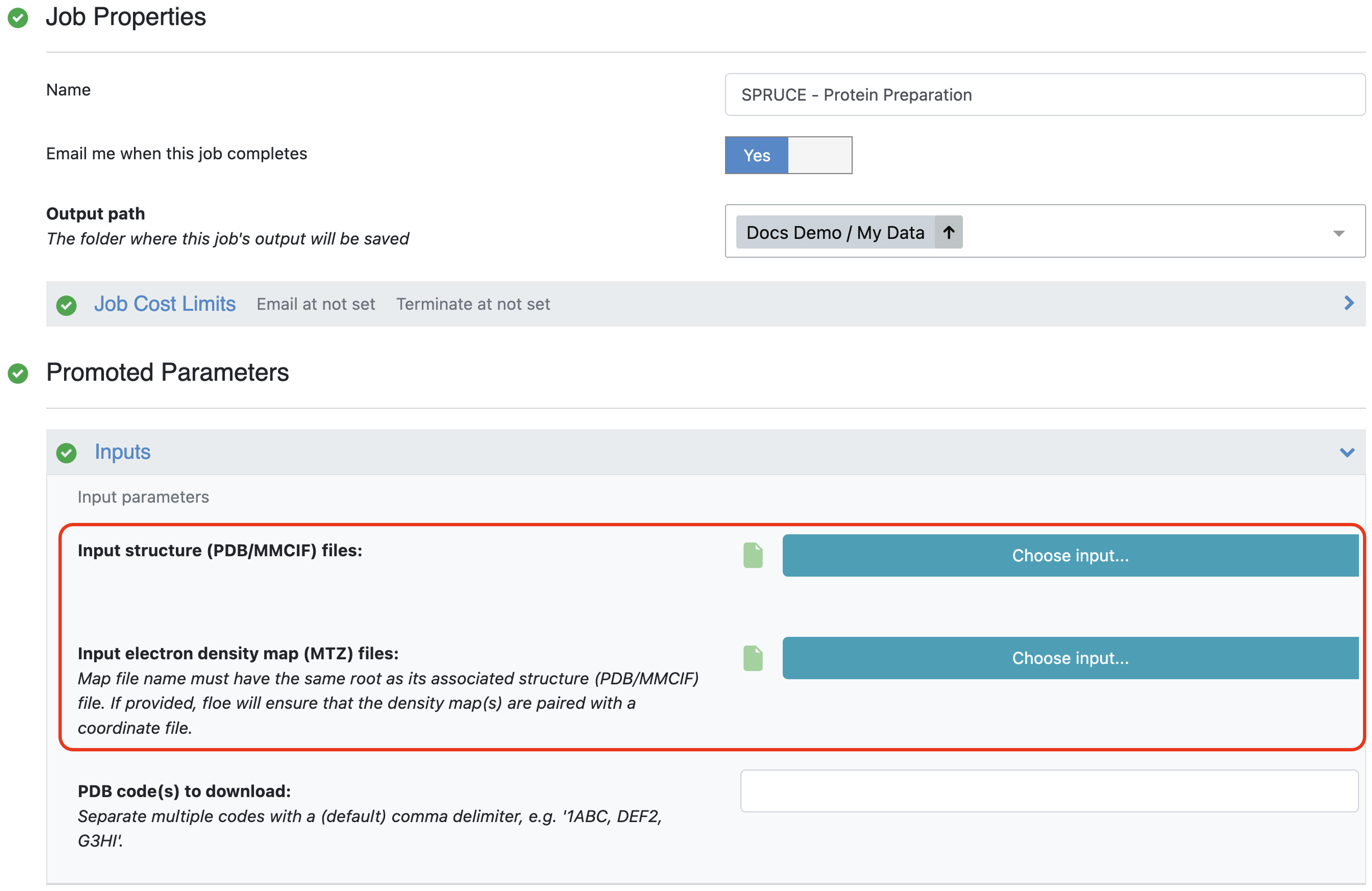
Figure 3. The area on the Job Form to provide PDB and MTZ files as input.
The files can be uploaded from a computer, pulled from a URL, or used from files already in Orion.
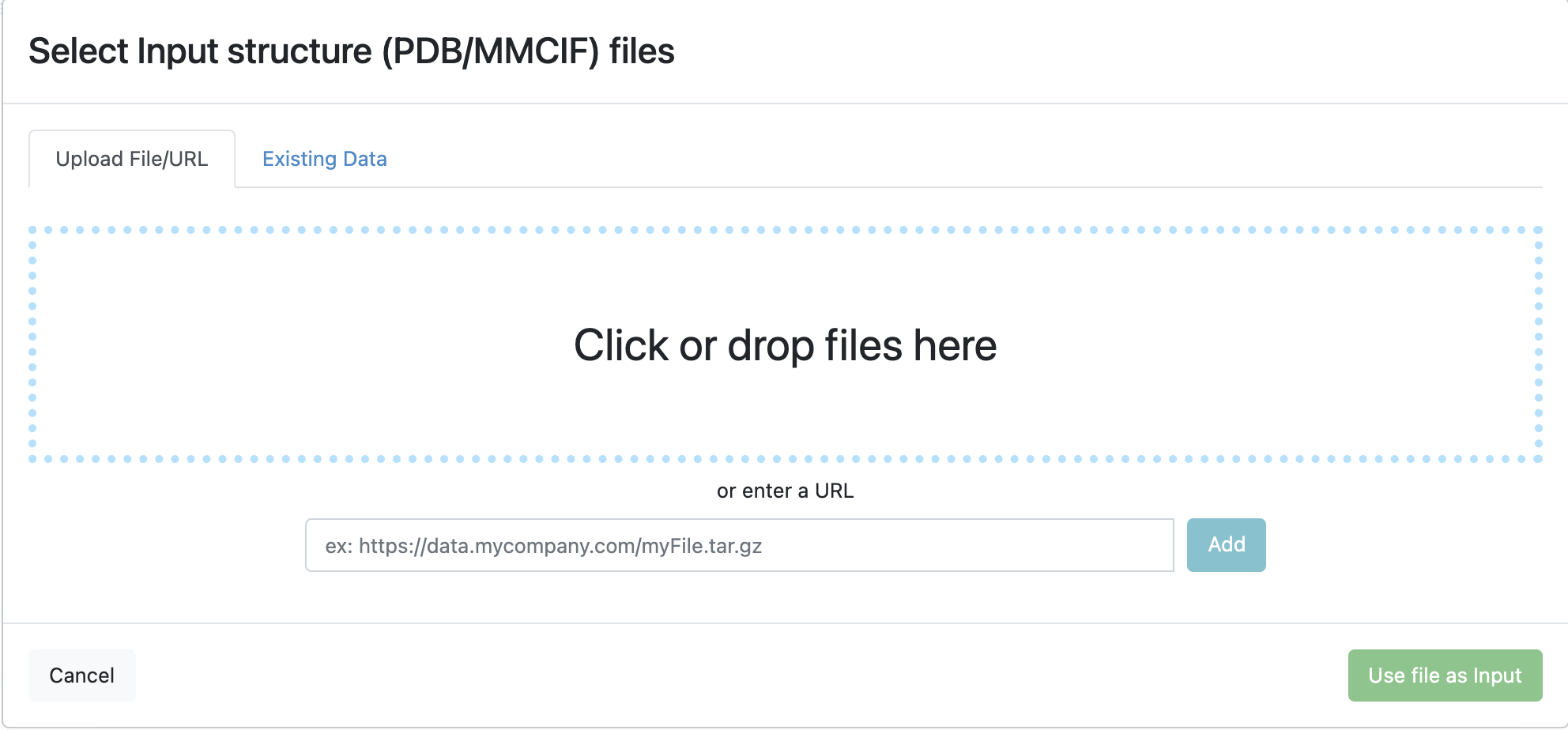
Figure 4. Where to upload files to Orion.
At least one protein structure must be input using either of the methods described above. The remaining inputs are optional and described in more detail in How To Guide.
Run the Protein Preparation Floes
An extracellular signal-regulated kinase 1/2 inhibitor (CDK2) is used in this tutorial to show how Spruce preparation can be done with or without a reference protein. When a reference structure is used, all submitted structures will be superposed, and the output design units will mirror the reference’s receptor. If no reference is used, all liganded design units will be output. Finally, if an apo structure is detected and no reference is given, pocket finding will generate design units based on structural pockets on the protein’s surface.
The most simple use is shown below using an experimental CDK2 structure (PDB code 5K4J) with only the
PDB code needed to run the
SPRUCE - Protein Preparation Floe.
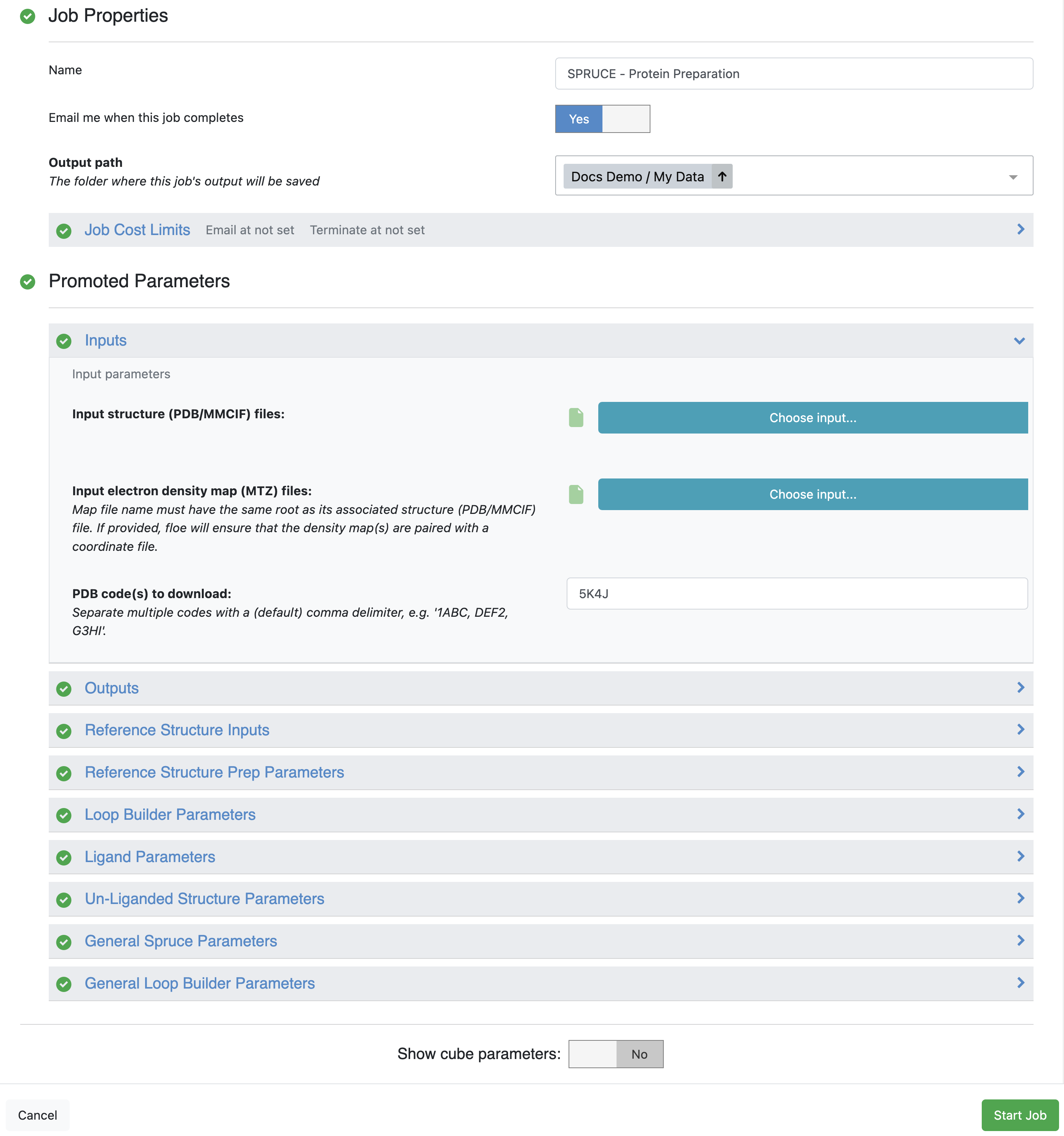
Figure 5. Parameters for the SPRUCE - Protein Preparation Floe.
The output dataset can be viewed in the 3D & Analyze page.
Run the Protein Preparation Floes Using a Reference Design Unit
Preparing a protein with a reference structure is similar to preparation without a reference structure. First, the protein structures to be prepared are input using any combination of the previously described input methods. For this example, we are going to prepare many related CDK2 proteins at the same time. The 5K4J structure is based on the P24941 UniprotKB ID; we can use this ID to search the PDB.
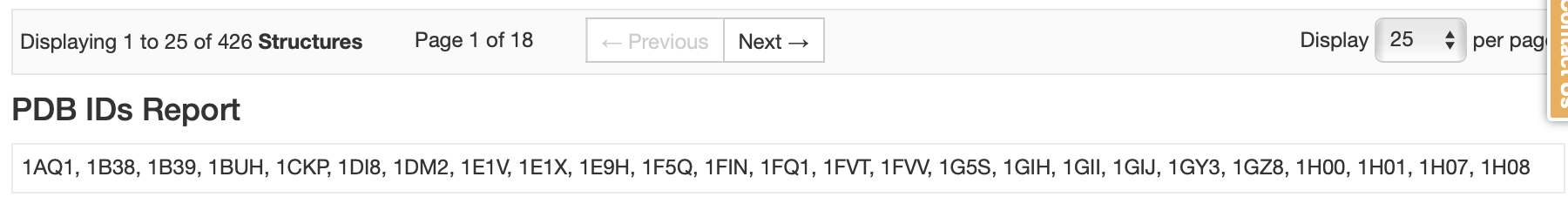
Figure 6. RCSB search results.
This yields a large list of PDB codes (with a tabular report showing the entry IDs). For this tutorial, we are going to prepare the first 25 hits with Spruce (1AQ1, 1B38, 1B39, 1BUH, 1CKP, 1DI8, 1DM2, 1E1V, 1E1X, 1E9H, 1F5Q, 1FIN, 1FQ1, 1FVT, 1FVV, 1G5S, 1GIH, 1GII, 1GIJ, 1GY3, 1GZ8, 1H00, 1H01, 1H07, 1H08). To identify the reference structure, use the Reference Structure Inputs parameters. Only one reference structure can be used per floe, and a reference structure can be identified using a dataset, PDB and MTZ files, or a PDB code. For this example, we use the CDK2 PDB code from above (5K4J) as the reference DU.
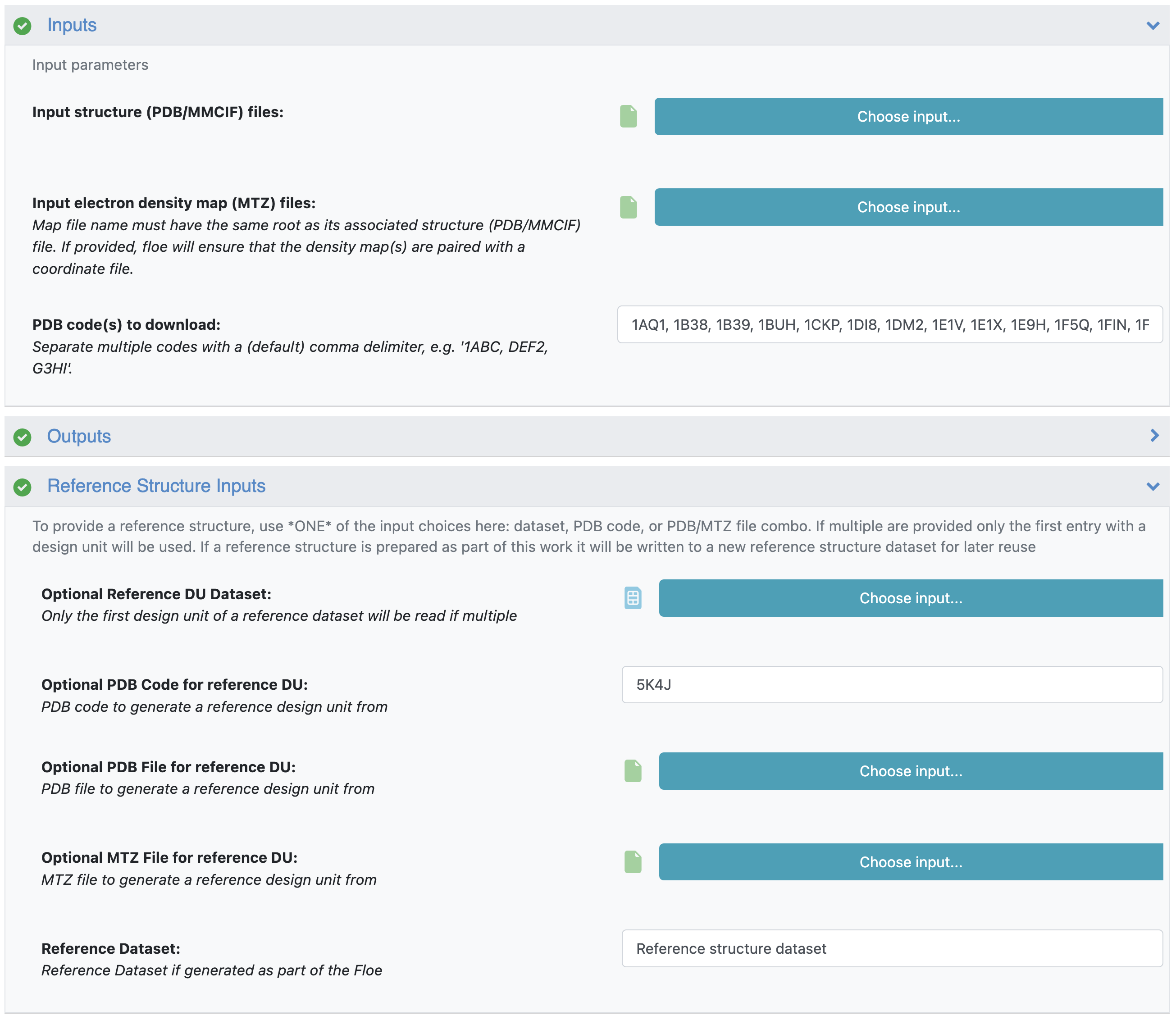
Figure 7. CDK2 subset preparation using a reference DU.
Reviewing the Floe Reports
This results in a dataset with 32 design units, and a dataset with three failed records. The failures are due to reference structure mismatches or inconsistencies in the structure and are filtered out with default settings. A dataset containing the reference structure is also created.
The 32 prepared design units from 24 PDB files are a result of multiple biological units generated from the crystal structure asymmetric unit, and certain experiments with alternate locations (configurations) near or in the binding site of interest. A Floe Report is also generated detailing issues with the prepared design units that may need further inspection. The issues may include partial residues where Spruce was unable to build a side chain, discussed in more detail in the theory documentation of Spruce TK.
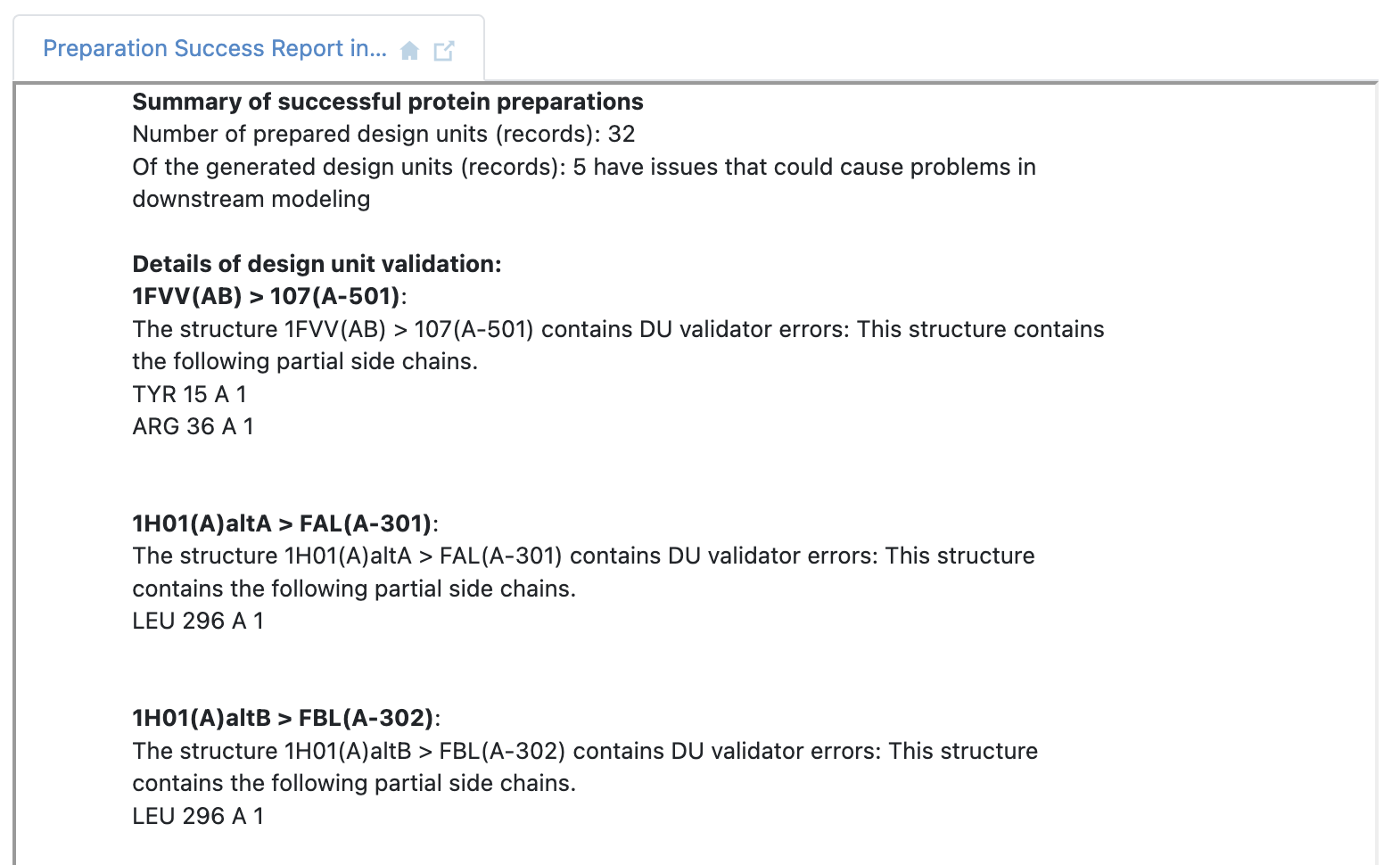
Figure 8. The Floe Report showing potential issues after structure preparation.
Analyzing the Results
First, we are going to mark both the reference DU dataset and the dataset containing the prepared DUs as active.
Figure 9. Activating datasets.
When analyzing a dataset containing a larger number of records, it can be helpful to view them in the 3D & Analyze page. This makes it easy to explore and filter on a property such as the Iridium classification. In this case, we have filtered out NA (Not Applicable: no Iridium Score could be calculated) and NT (Not Trustworthy) structures. Furthermore, the remaining structures have been sorted on the classification. Filtering can be done here because we can also sort the records based on the Iridium classification, and we can inspect the various depiction images, giving us a high level overview of the dataset before switching to the 3D view.
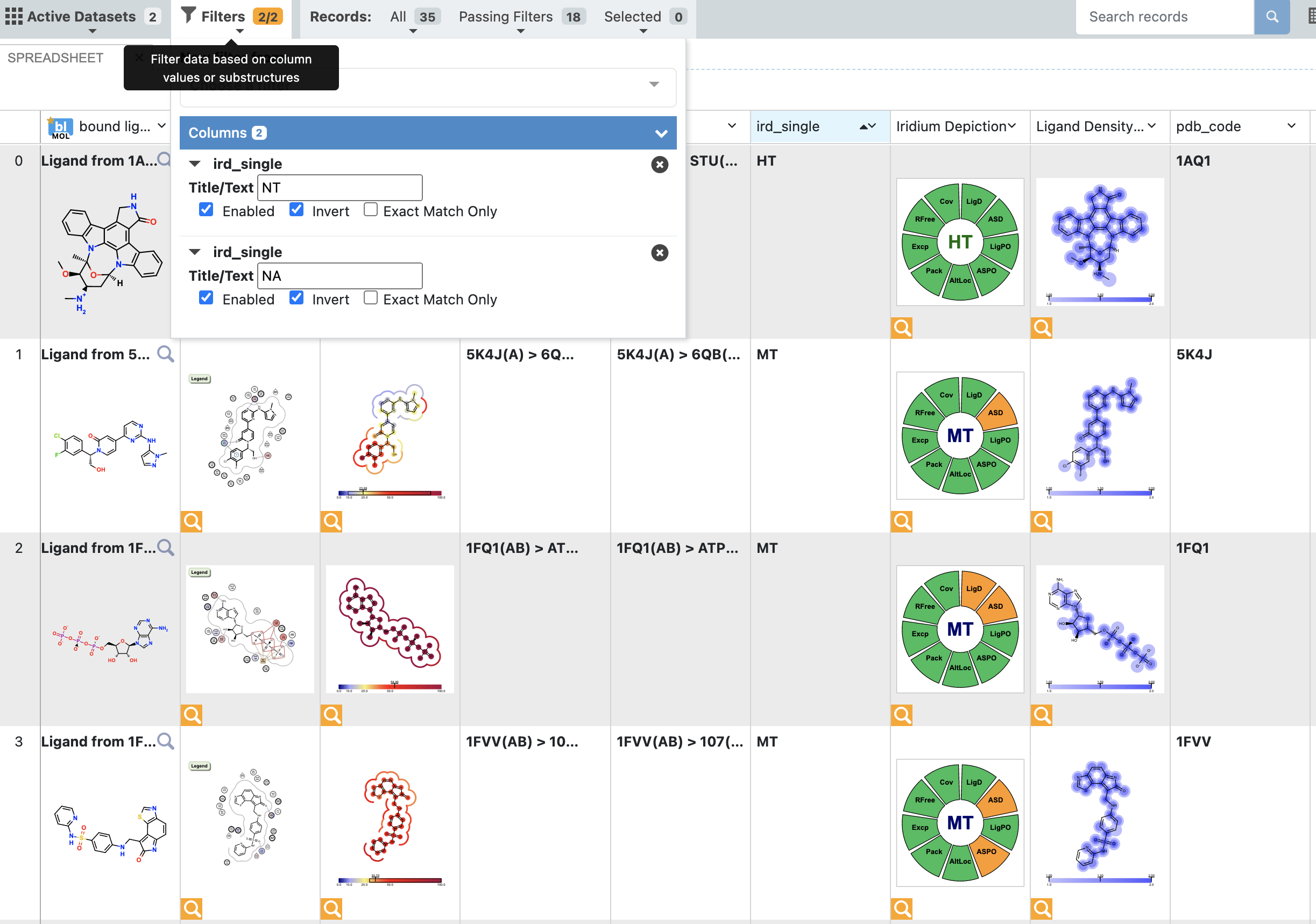
Figure 10. Filtering a dataset.
Once filtering has been applied, the relevant structures can be viewed in the 3D Viewer. Since a reference structure was used, all the structures are superposed, allowing for easy comparison and exploration of differences in ligand binding poses or binding site conformations. In Figure 11 below, the reference structure dataset is shown with protein-ligand interactions indicated.
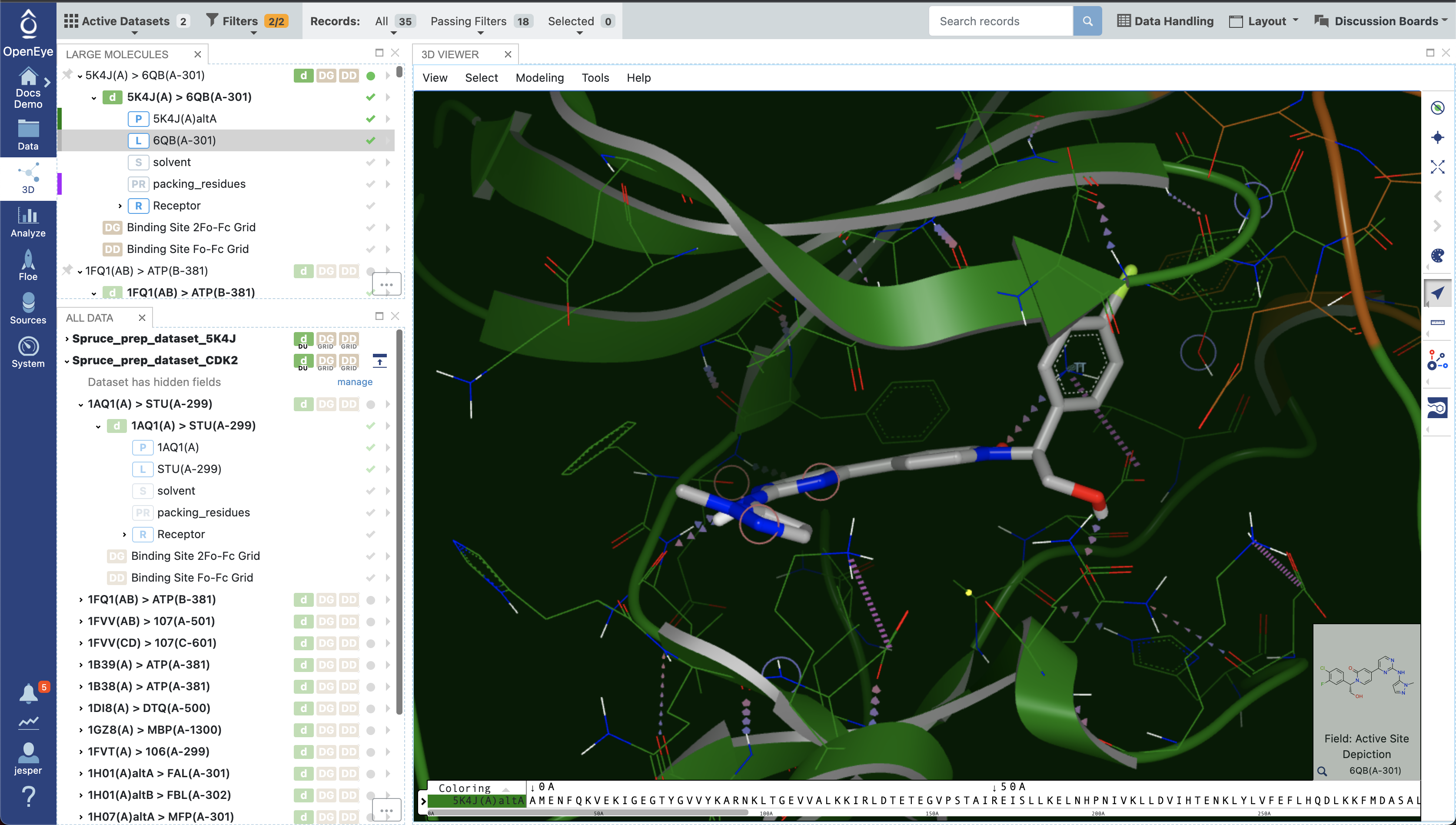
Figure 11. The design unit in the 3D Viewer.
Data stored on the DU record makes it possible to inspect the experimental electron density maps with just a few mouse clicks.
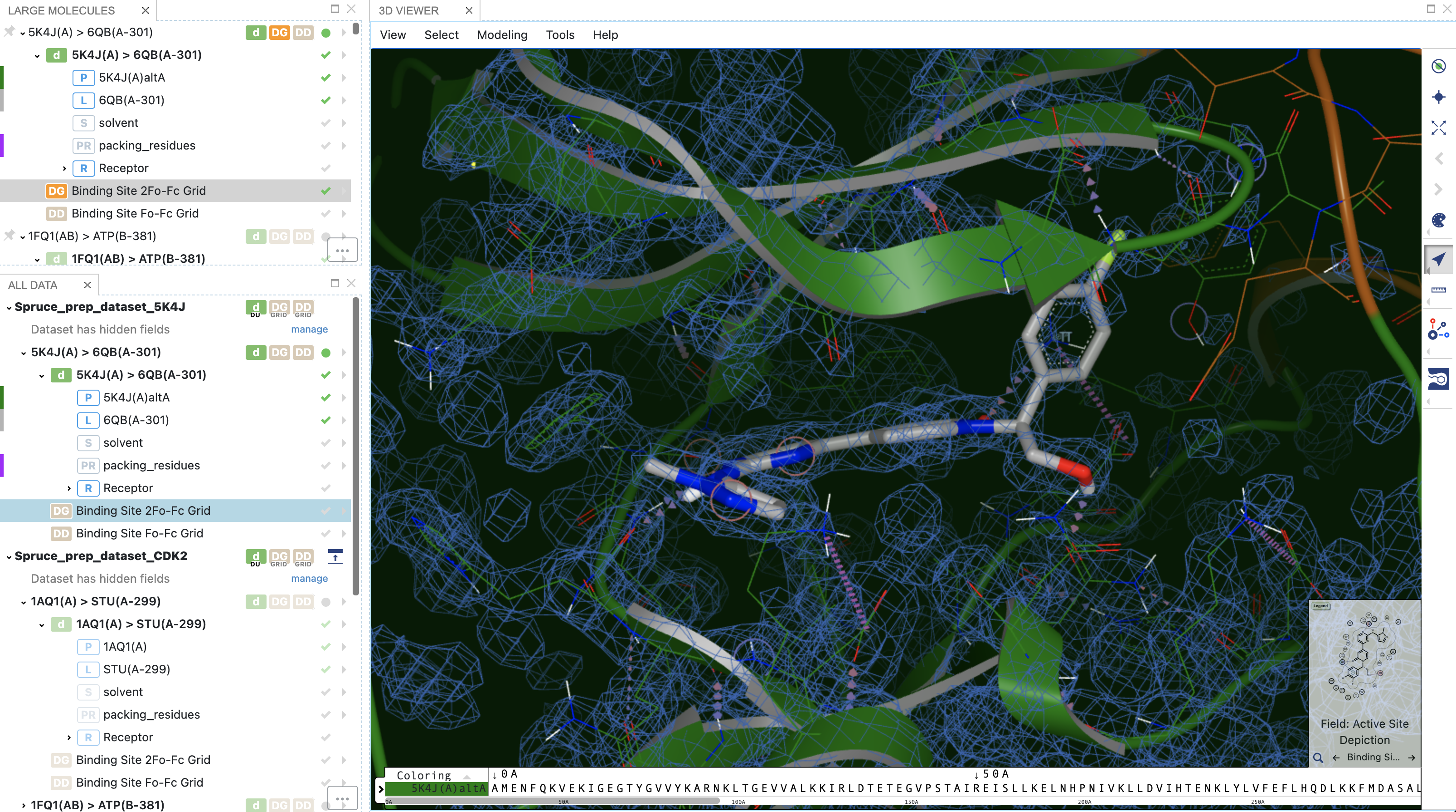
Figure 12. 2Fo-Fc electron density grids shown with the structure.
Multiple structures may be displayed at the same time, using different color schemes for the bound ligands. Inspecting differences in binding poses, particularly specific binding interactions such as hydrogen bonds, salt bridges, and pi-type interactions, can be helpful in explaining experimental data, such as differences in activities or differences in the specificity of a compound between different targets.

Figure 13. Superposed design units in a dataset with a reference DU. One of the bound ligands is shown with a different color scheme.
Depending on the subsequent modeling task, one or more design units can be selected and saved as a new and curated dataset. In this case, we could save only the structures with an HT (Highly Trustworthy) Iridium classification. It is important to note that many MT structures are also relevant for use in modeling tasks. The MT classification indicates that a structure should be inspected closely prior to being used in computations, in particular the ligand binding sites, to avoid unexpected results.
Calculation of the Iridium classification requires electron density maps to be supplied in MTZ format. If these maps are not available, the Iridium classification cannot be calculated, and classification is marked as NA. Furthermore, Iridium cannot be calculated for apo structures, which are also marked NA.
Next Steps
Design units prepared by Spruce can be used for a variety of modeling tasks. One integral part of the DU preparation process is the generation of a receptor binding site (docking grid), which allows the DU to be used for ligand docking or posing tasks.