Antibody MD Simulations Tutorial
Antibody Molecular Dynamics Simulations
Molecular dynamics (MD) simulations can be run on the generated models using the Antibody MD Simulations with Conformational Analysis Floe. Along with providing the tools to quickly run MD simulations on target antibodies, the floe also identifies the different possible conformations for the target antibodies by clustering the obtained frames. This is not only useful for determining the different binding modes for the target antibodies, but also for analyzing the conformations possible for the hypervariable complementarity-determining regions (CDR) on the antibodies.
Multiple structures to be run separately can be provided as a dataset. These structures can only be outputs of the Antibody Experimental Structure Prep and the Antibody Sequences to 3D Models Floes. Any custom structure or PDB structure needs to be passed to the Antibody Experimental Structure Prep Floe before being used for MD simulations.
Parameters such as the ligand force field, protein force field, trajectory interval, and more can be modified in the MD setup section on the Job Form.
Along with the frames from the simulation, the floe outputs a dataset of the cluster centroids representing the different conformations of the antibody. These are determined by calculating a pairwise RMSD matrix for all the frames and then using HDBSCAN.
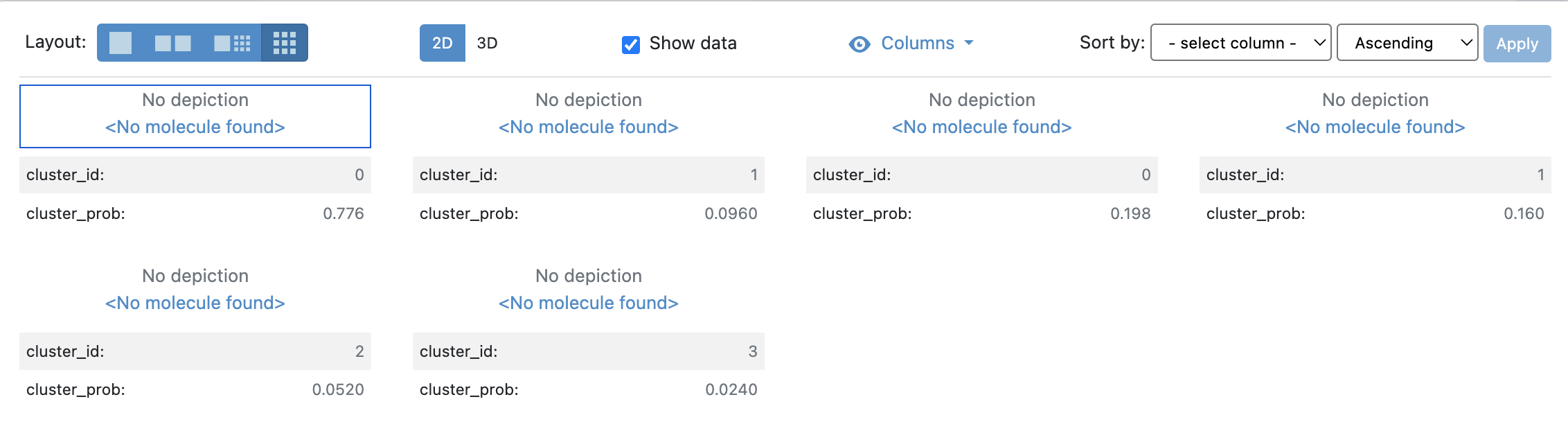
Figure 1. Centroid dataset generated by conformational analysis of the MD simulation results for 5FUU.
The centroids can also be visualized in the 3D Viewer for structural comparison.
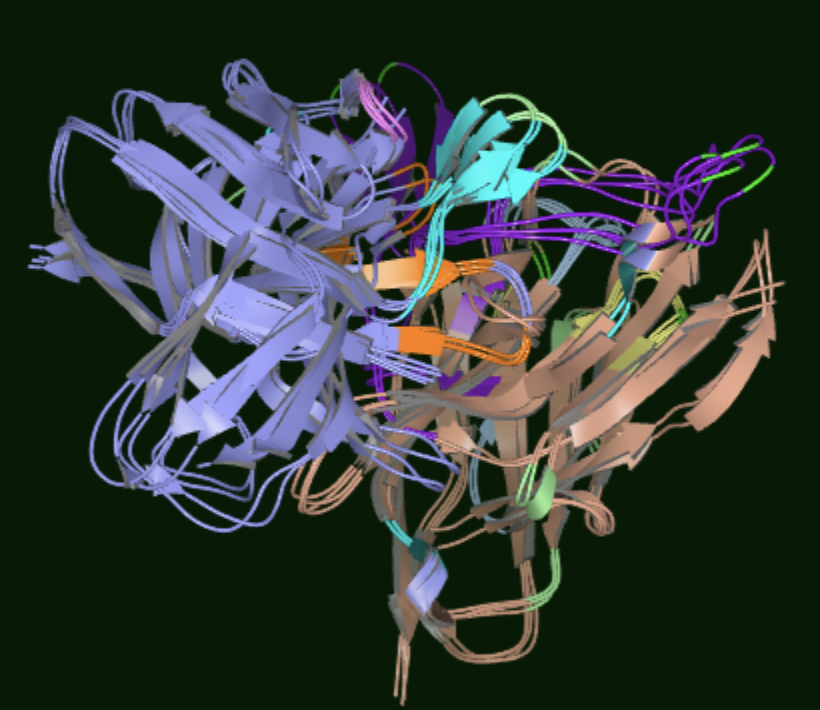
Figure 2. Centroid structures representing the different conformations for 5FUU.