3D Antibody Modeling Tutorial
The antibody modeling floes can be used to generate models for target antibodies and analyze them. Antibody model generation floes rely on ImmuneBuilder to build models and use SPRUCE to prepare them for downstream modeling applications. This tutorial explains how to import sequence data and generate antibody models, as well as how to prepare experimental structures for downstream modeling. Other features of the package can be found in the floe reference documentation.
Floes Used in This Tutorial
The floes used in this tutorial are:
Importing Sequences
Antibody sequences can be imported into Orion in the following two ways.
Both methods convert the information into a dataset that can then be used with the Antibody Sequences to 3D Models Floe.
In the first method, a CSV file containing a row for each antibody of interest should be provided, which will be automatically converted to a dataset by Orion when uploaded. The file should contain a header with the column contents. Each row should contain a field for a unique identifier, a field for the heavy chain sequence, and a field for the light chain sequence.
The second method requires providing a FASTA file in the appropriate format to the Import Antibody FASTA Files Floe. Multiple Fv fragments can be provided in the FASTA file as long as each entry conforms to the required format. A template for FASTA entries is shown below.
>Bevacizumab_H
EVQLVESGGGLVQPGGSLRLSCAASGYTFTNYGMNWVRQAPGKGLEWVGWINTYTGEPTYAADFKRRFTFSLDTS
KSTAYLQMNSLRAEDTAVYYCAKYPHYYGSSHWYFDVWGQGTLVTVSSASTKGPSVFPLAPSSKSTSGGTAALGC
LVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKS
CDKTHT
>Bevacizumab_L
DIQMTQSPSSLSASVGDRVTITCSASQDISNYLNWYQQKPGKAPKVLIYFTSSLHSGVPSRFSGSGSGTDFTLTI
SSLQPEDFATYYCQQYSTVPWTFGQGTKVEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKV
DNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
Each entry must have two associated sequences: one for the heavy chain indicated by “H” following the antibody ID, and one for the light chain indicated by “L” following the antibody ID. The antibody ID should not contain any underscores. This FASTA file can then be provided to the Import Antibody FASTA Files Floe to generate an antibody sequence dataset.
Antibody Model Generation
Antibody models can be generated either from sequences of the heavy and light chains or from their experimentally determined structures.
Starting with Sequences
The Antibody Sequences to 3D Models Floe uses machine learning models to generate models from sequences. Sequences in a dataset are provided to generate models for antibodies. The method requires a dataset which contains a unique identifier, the heavy chain sequence, and the light chain sequence of each antibody of interest. This information must be provided to the Antibody Name Field, VH Sequence, and VL Sequence parameters, respectively. This dataset can be generated using the methods discussed above.
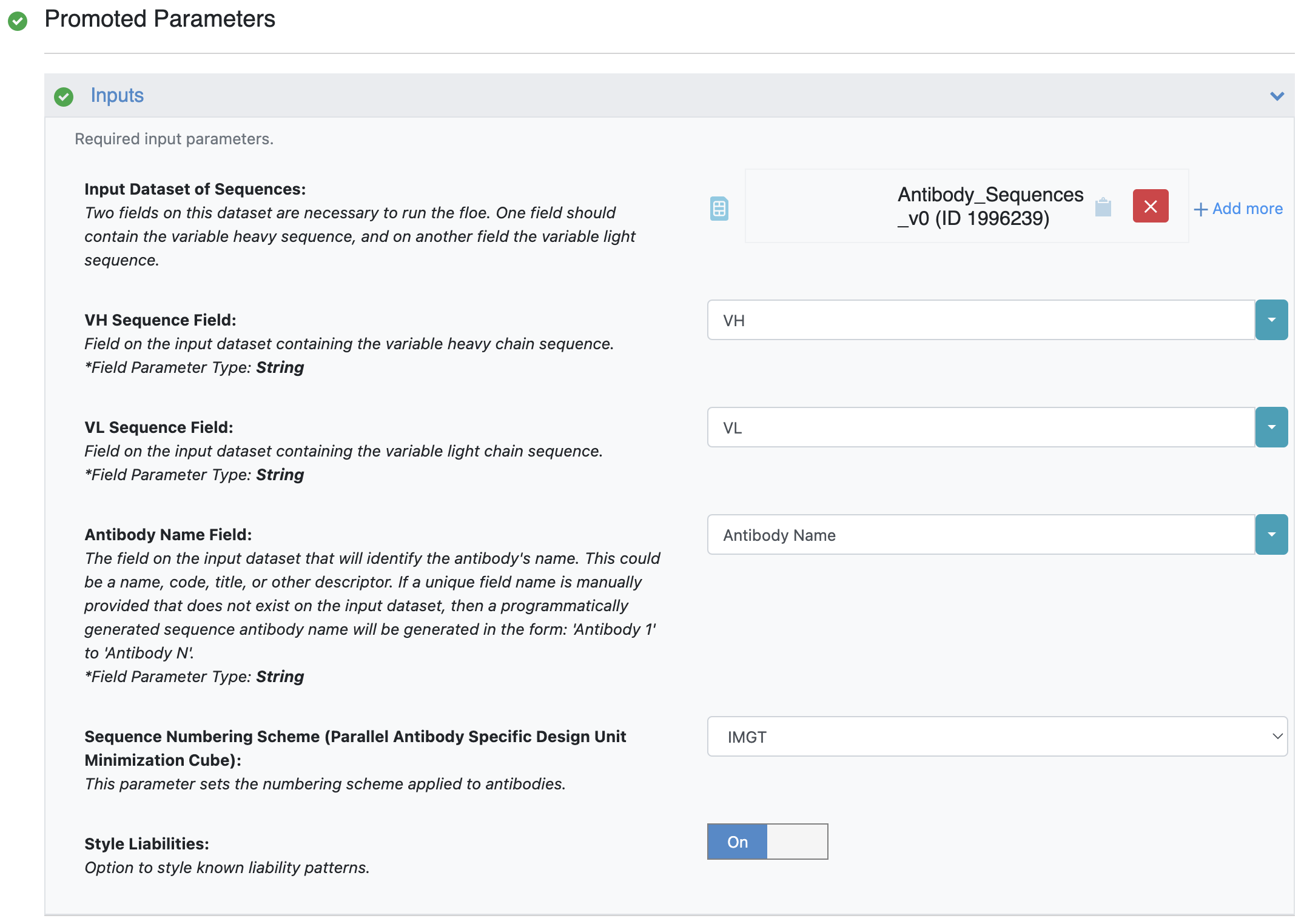
Figure 1. Providing sequences as a dataset.
Starting with Experimental Structures
The Antibody Experimental Structure Prep Floe can be provided structures or PDB codes of antibodies to generate models useful for downstream applications. This is important, as experimental structures by themselves cannot always be used by other tools and have to be prepared first. Moreover, this floe can add appropriate annotations to the antibody Fv region, fill in missing loops, and analyze the surface.
Structures can be provided to the floe using the following two methods.
Choosing or uploading the structure file
Providing PDB codes for the relevant structures
The first method requires the structure file to be in PDB or MMCIF format. It is also possible to provide an electron density map (MTZ) file with the associated structure file if it is available.
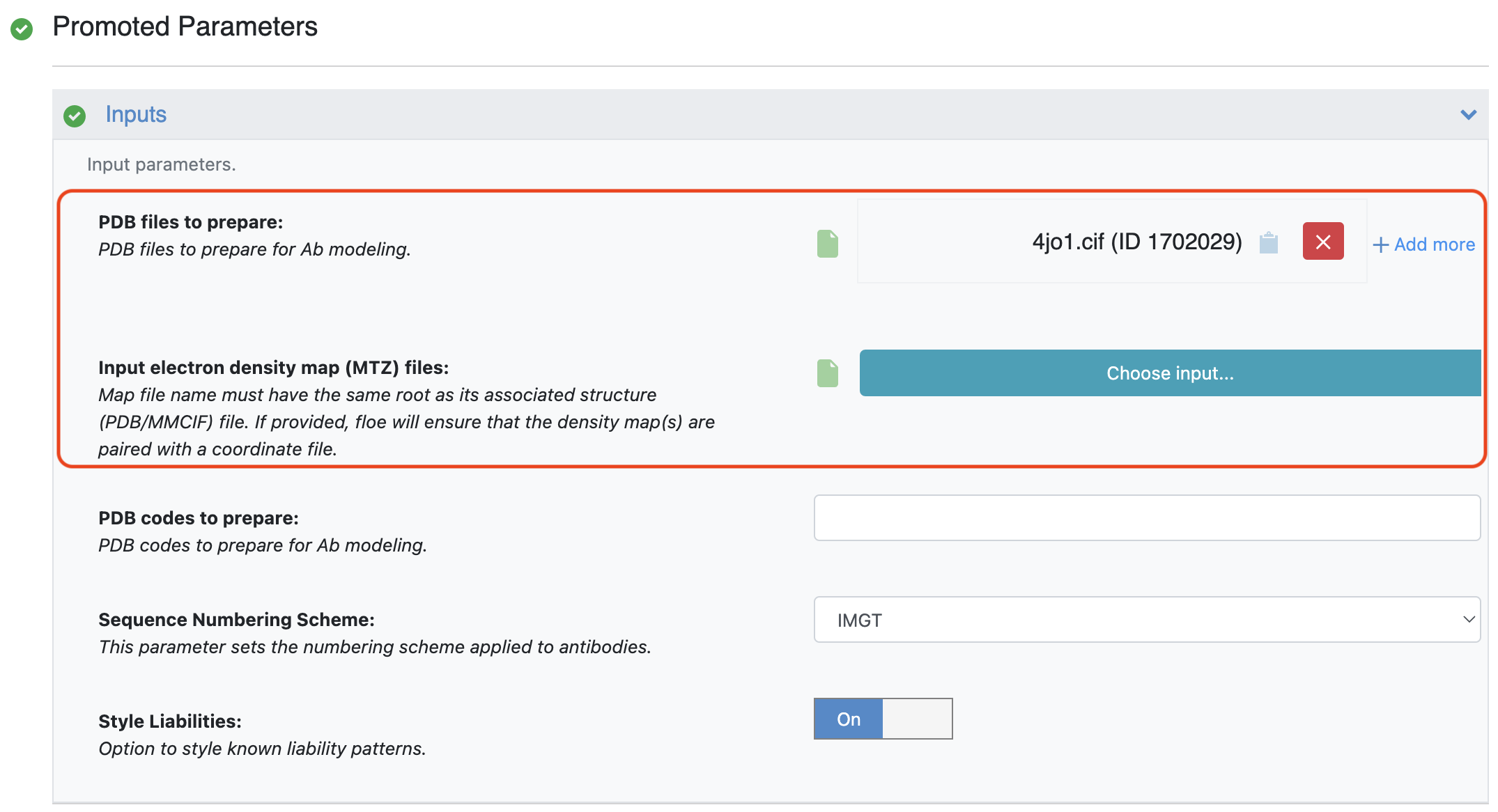
Figure 2. Providing structure and electron density map files. Electron density map files are optional.
The second method requires entering the relevant structure PDB codes into the PDB Codes to Prepare parameter. The PDB codes should be separated by commas.
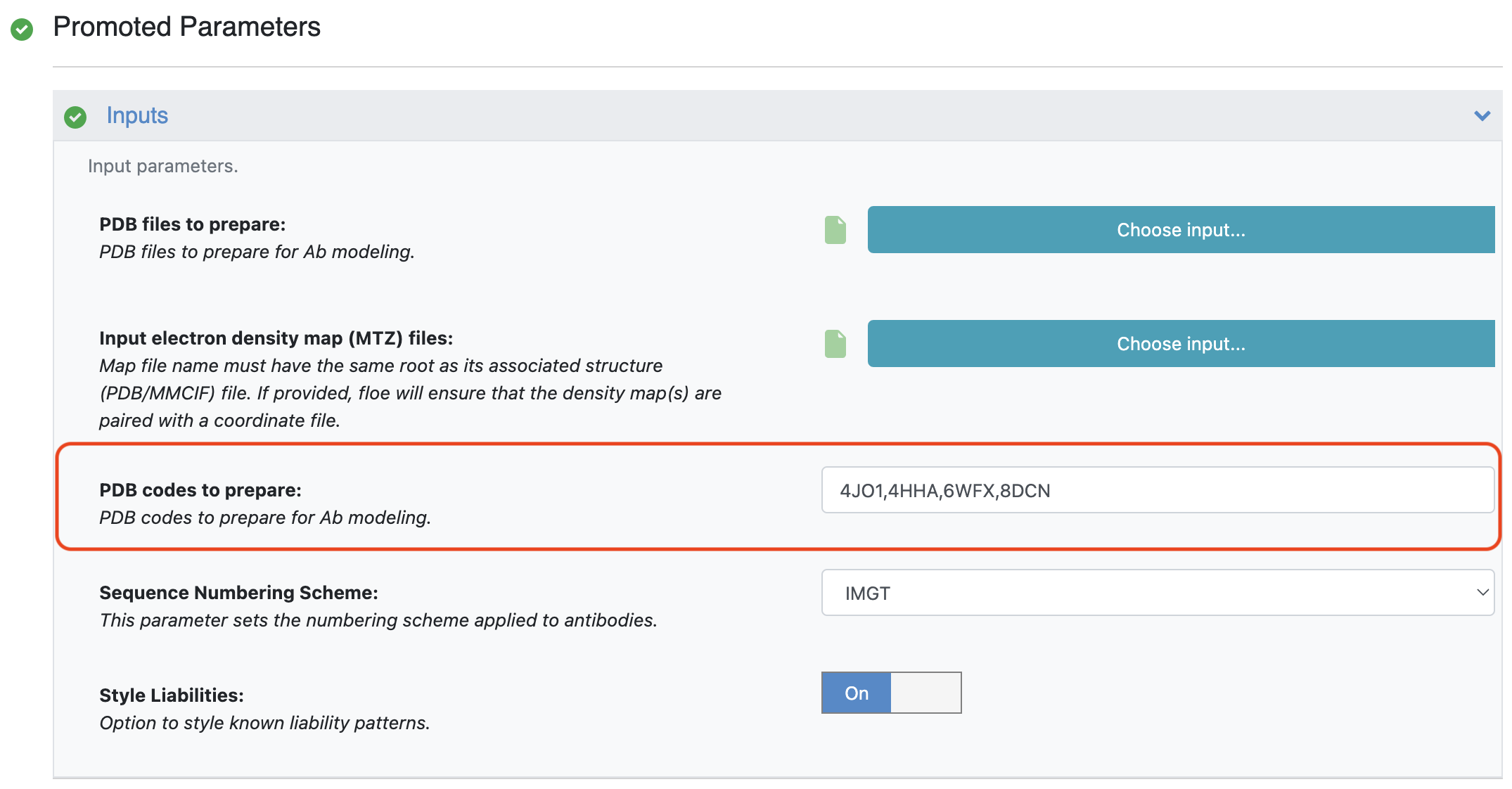
Figure 3. Providing PDB code(s).
Once the structures have been provided, loop builder parameters can be set to build missing loops and to build missing tails.
Modeling Results
The results of these floes can be visualized in the 3D Viewer. By default, these floes also add surface patches to the models and convey information about surface properties. More information on the surface patches can be found in the Understanding Patches tutorial.
The modeling results for these antibodies also provide metrics for developability based on guidelines from therapeutic antibody profiling [Raybould-2019]. They consist of the following metrics.
Total CDR length
Patches of Surface Hydrophobicity (PSH), CDR vicinity
Patches of Positive Charge (PPC), CDR vicinity
Patches of Negative Charge (PNC), PNC, CDR vicinity
Structural Fv Charge Symmetry Parameter (SFvCSP)
Based on the thresholds defined by Raybould et al. counts of amber and red flags are added to the modeling results to highlight the developability risk.