Deciding on Clustering Method
The clustering floes in the Small Molecule Modeling package provide many options for clustering molecules based on their similarity score. This section provides basic guidance on which clustering floe to use based on your application and input data. For brief overviews of each clustering method, and links to specific tutorials, see the methods section.
Floes Used in This Tutorial
The floes referenced in this tutorial are:
[Deprecated] DBSCAN 2D Similarity Clustering
[Deprecated] DBSCAN 3D Similarity Clustering
[Deprecated] Hierarchical 2D Similarity Clustering
[Deprecated] Hierarchical 3D Similarity Clustering
[Deprecated] K-Medoids 2D Similarity Clustering
[Deprecated] K-Medoids 3D Similarity Clustering
Decision Tree
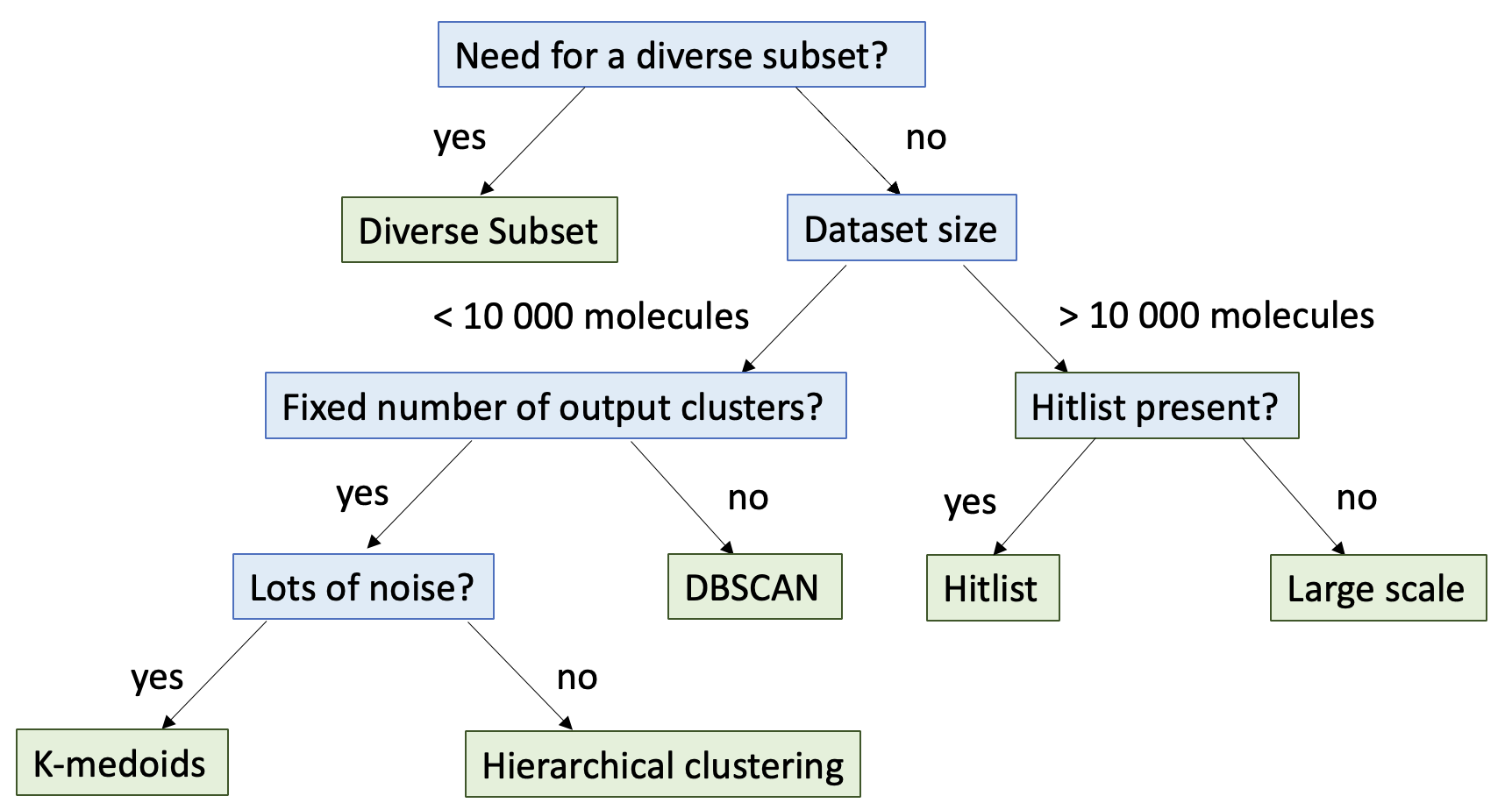
Size Constraints
Is your dataset larger than 10,000 molecules? If so, you are likely to get results much faster using the large scale clustering floes. If the dataset is larger than 30,000 molecules, it will fail the small scale clustering floes.
For datasets smaller than 10,000 molecules, refer to the applications section to decide clustering based on a specific application, or the small scale clustering section for other selection criteria. Please keep in mind that these are only general guidelines for choosing clustering method. It is recommended to consult academic literature to determine the best method for your use case.
Application-Based Clustering
Do you have a specific application in mind for clustering?
Clustering a hit list: use a Hit List Clustering Floe, as described in the Hitlist Clustering tutorial.
Selecting a specific number of clusters: do you need only the subset?
If yes, use the 2D or 3D Diverse Subset floe.
If no, use the K-Medoids or Hierarchical clustering methods.
Small Scale Clustering
Do you want a specific number of clusters?
If yes, you will need to use the Hierarchical clustering or K-medoids methods.
If no, and you don’t want to guess the appropriate number of clusters, you will need to use the DBSCAN method.
Does your dataset have a large number of outliers or lots of noise?
If yes, the DBSCAN and K-medoids clustering methods are less sensitive to outliers. Also, if your dataset has non-globular clusters or clusters of multiple sizes, the DBSCAN method is more likely to be effective than the K-medoids clustering method.
Do you want to have better control over the size of the largest cluster?
If yes, the DBSCAN clustering method allows you to choose a minimum or maximum size of the largest cluster.
Clustering Methods Overview
Note
The DBSCAN, K-Medoids, and Hierarchical Clustering Floes have been deprecated. For ease of use, the clustering methods from these floes have been incorporated into the Clustering: 2D Fingerprint, up to 100K compounds and Clustering: 3D Shape/Color, ~10K compounds Floes.
DBSCAN Clustering
DBSCAN clustering uses scikit-learn’s DBSCAN method. DBSCAN stands for Density-Based Spatial Clustering of Applications with Noise. Unlike K-medoids or hierarchical clustering, DBSCAN views clusters as areas of high density separated by areas of low density. For more details, refer to the scikit-learn documentation.
K-Medoids Clustering
K-medoids clustering uses scikit-learn’s extra K-medoids method. K-medoids tries to minimize the sum of distances between each point and the medoid of its cluster. Its principal advantages are that it can produce the exact number of requested clusters, and that it is less sensitive to outliers than other methods of clustering, including k-means and hierarchical clustering. For more details, refer to the scikit-learn documentation on k-medoids.
Hierarchical Clustering
Hierarchical clustering uses scikit-learn’s AgglomerativeClustering method to cluster using a bottom-up approach. Each member starts in its own cluster, and clusters are merged together in a series of merge steps that occur according to the linkage criteria that is selected. More details can be found in the scikit-learn documentation.
Hit List Clustering
The 2D or 3D Hitlist Clustering Floes take a large scale input dataset and require a float score field on each record. That score field will be used to direct clustering, and also be used to rank clusters in the clustering report, and provide information about average or best rank in each cluster. For more details, see the hit list clustering tutorial.
Diverse Subset Floes
The 2D or 3D Diverse Subset Floes take an input dataset, and select N members from that dataset with clustering, using sphere exclusion for large datasets, and K-medoids for small-scale clustering. For large datasets, large scale diverse subset may be significantly faster than clustering itself. For more details, see the diverse subset tutorial.
Large Scale Clustering
With a large dataset, you have effectively three options for 2D or 3D clustering:
Do you only need a diverse subset? Use the 2D or 3D Diverse Subset Floes.
Do you have a hit list or another type of dataset that has a score for each molecule? Use the 2D or 3D Hitlist Clustering Floes or the Clustering: 2D Scaffold Floe.
If the answer to 1 and 2 is no, use the 2D or 3D Large Scale Similarity Clustering Floes, or the Clustering: 2D Fingerprint, up to 100K compounds and Clustering: 3D Shape/Color, ~10K compounds Floes.
The 2D and 3D Large Scale Similarity Clustering Floes use the same method (directed sphere exclusion) as the Diverse Subset and the Hitlist Clustering floes. Unlike the Diverse Subset Floes, they also output cluster members. Unlike the Hitlist Clustering Floes, they don’t require a score field to be used in clustering, or sorting output clusters. For more details, see the large 2D clustering tutorial.
The Clustering: 2D Scaffold (~25M compounds) Floe is discussed in more detail in the How-to Guide for Molecular Clustering.
The Gigadock Floe in the Large-Scale Floes package can also be used for clustering. For more information, see the Cluster Hit List Poses from Gigadock tutorial.