Tutorial: Focused Library – Molecule Input Floe with BlockBuster and XLogP Filtering for Postprocessing
The Focused Library – Molecule Input Floe is another floe that can be used for generative design. The input is a molecule with no attachment site specified. This floe detects synthetic handles and enumerates products using compatible reagents as defined by the user. This tutorial will use the database prepared in the Create a Reaction and Reagent Database Floe tutorial. If you want to go through this section without preparing your own database, you can use the OpenEye databases provided in the Organization Data folder.
This tutorial uses the following floe:
Focused Library – Molecule Input Floe
Enumerate Reagents for an Input Molecule Using a Built Database
In your tutorial project, go to the Data page. Click the “Add Data” button and then choose Sketch. Paste the SMILES string c1cc(ccc1C(=O)O)S(=O)(=O)Nc2ccc(c(c2)C#N)Oc3ccc(c(c3)C(F)(F)F)Cl into the Sketcher.
Add the name lead_molecule to the sketched molecule, click the “Save” button, and press “Done” (Figure 1). A pop-up window will ask to save the molecule as a new dataset. To do so, choose a name and click “Save” in the pop-up window.
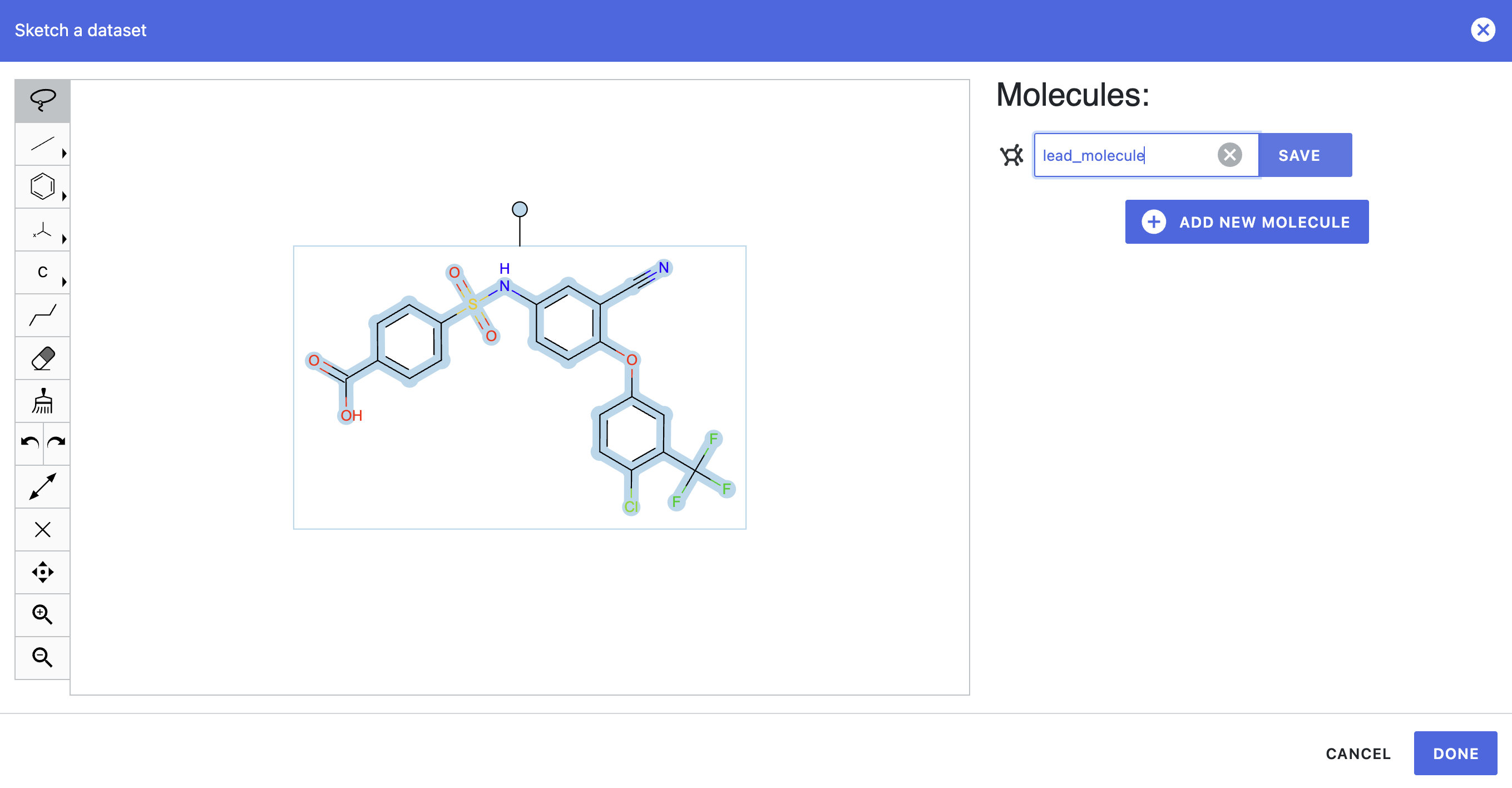
Figure 1. Input molecule passed to the Sketcher.
On the Floe page, choose the Focused Library – Molecule Input Floe from the OpenEye Generative Design Floes – Advanced Package. Click “Launch Floe” to bring up the Job Form.
Inputs
Lead Molecule Dataset (Lead Molecule): Choose the dataset lead_molecule you just created.
Reaction & Reagent Database (Classify Molecules): Select the Mcule reaction and reagent database that you created in the Create and Inspect a Reaction and Reagent Database from a SMILES File tutorial.
Output Dataset: Title this as Reaction_products_tutorial.
Focused Library Options
Reactions or Reagents: Choose Schotten-Baumann_amide:Carboxylic_acids. The functional group described along the reaction specification refers to the functional group that must be present in the input query molecule.
Focused Library Filtering Options
Filter Output: Toggle to On. This will ensure that all generated molecules will pass through the Blockbuster filter before being written to the output file. For more information about OpenEye default filters, please click here.
Reagent Processing Options
Allow Functional Group Conversions: Turn this toggle Off to allow only reagents present in the database to be enumerated. In this tutorial, we will enumerate all the reagents in the database that contain an amine able to undergo a Schotten-Baumann reaction with the lead molecule.
Maximum Reagents: Leave this field blank to allow for complete sampling.
Sample Reagents: Set this parameter to 100 to sample 100% of the total available reagents (Figure 2).
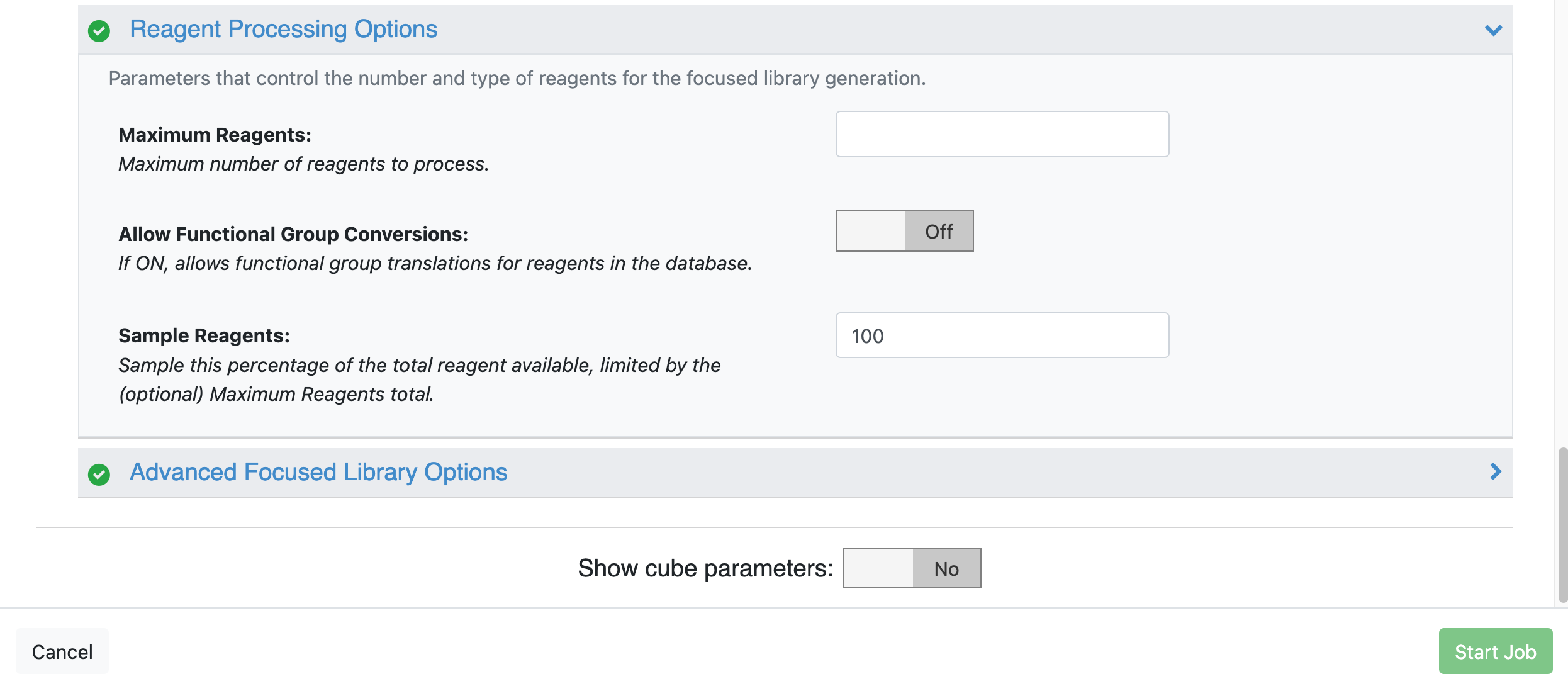
Figure 2. Reagent Processing Options input parameters from the Focused Library – Molecule Input Floe.
Advanced Focused Library Options
Strict Classification: Toggle this Off. When this parameter is Off, the scaffold will not be validated for the detection of prohibited functional groups in the lead molecule. Only the required functional group (carboxylic acid, in this case) will be considered. If this is On, the floe will fail because of the presence of the aryl chloride. Allowed and prohibited functionalities can be viewed in the Sample Reaction Classification file. If you would like to verify the difference, you can repeat this tutorial with this toggle On and with the chloride removed from the lead molecule.
Click “Start Job” to begin the floe.
Visualize and Filter Enumerated Products on the 3D & Analyze Page
Once the job is complete, activate the generated dataset and load it in the 3D & Analyze page.
Select ‘Filters’ from the Active Data Bar at the top of the page. Click on the “Choose a Filter” box and select XLogP, with a range from 3 to 5. Ensure that “Enable” is checked and press Enter on your keyboard to set the range (Figure 3).
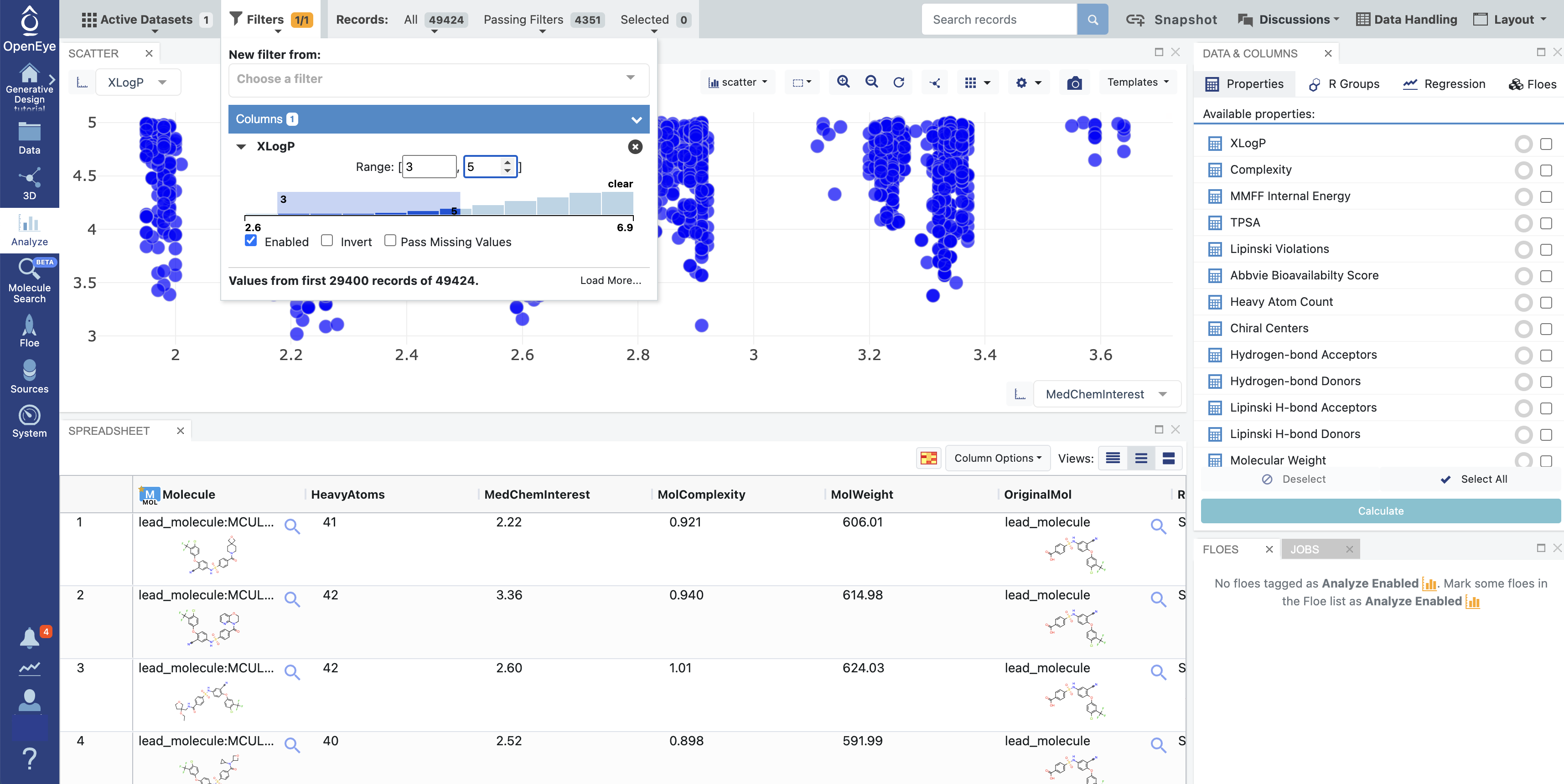
Figure 3. 3D & Analyze page showing the XLogP filter.
On the Active Data Bar, select Save Records under the ‘Passing Filters’ drop-down. Select New Dataset as the destination and enter the name Filtered_Schotten-Baumann_amides. Next, choose the “Replace Active Data” radio button and click the “Save” button. The Analyze page will now display your filtered dataset, complete with identifiers for the building blocks that were used to build the compounds of interest should you want to order starting materials for a particular structure from a vendor.