Tutorial: Search for Similar Binding Sites with SiteHopper
SiteHopper is a tool to superpose and compare protein binding sites. The input is a protein–ligand (or apo) complex where the binding site is converted to a searchable site called a patch. SiteHopper can efficiently compare this patch to other binding site patches, which are stored in a pre-generated database.
A search for similar binding sites can be useful for a number of different reasons, two of which are:
A similar binding site in a related or unrelated protein can indicate possible off-target risks. Finding and comparing these binding sites can be helpful for improving compound selectivity.
A ligand from a similar binding site can potentially be used as a tool compound or as an early stage hit for a target. Especially interesting are results where the binding site similarity is high while the ligand similarity is low: think of this as scaffold hopping based on binding site similarity.
In this tutorial, a dataset of binding site patches will be searched with a query structure from the CDK2 target using the SiteHopper Search Floe. The results can be visualized in both the Analyze page and the 3D Viewer. The patch database preparation should only be run once and is afterwards shareable with others; we have shipped a few large searchable databases as well.
This tutorial uses the following floes:
Create a Tutorial Project
Note
If you have already created a tutorial project, you can reuse the existing one.
Log into Orion and click the “Project” button on the blue navigation bar. Then click on the “Create Project” button in the upper right, and in the pop-up window, enter Tutorial for the name of the project and click “Create Project” in the window.
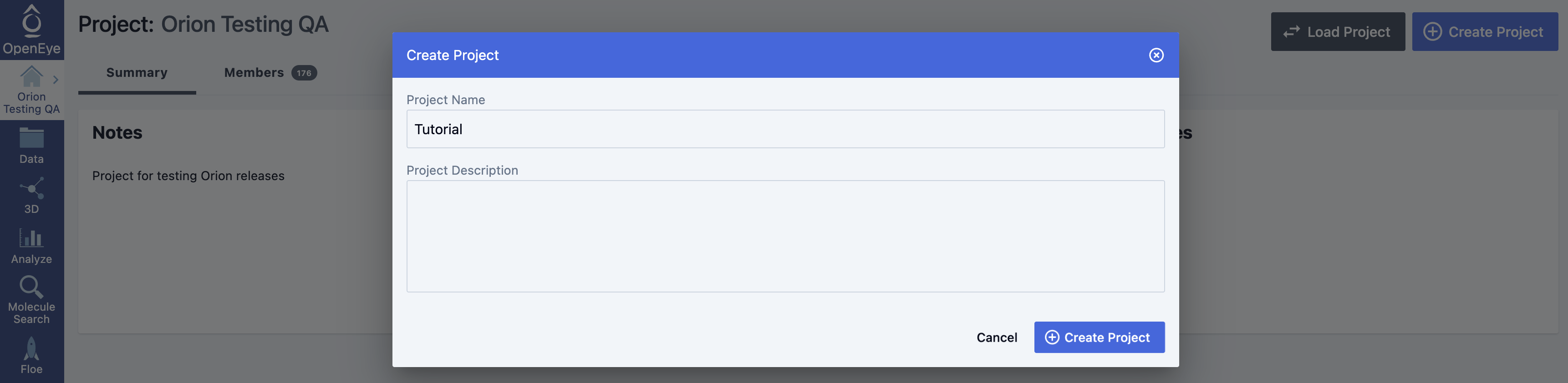
Figure 1. How to create a project.
Prepare Patch Dataset
Note
If you have already created a patch collection, you can reuse it for search. OpenEye offers pre-generated collections that use design units in MMDS that are converted into SiteHopper searchable collections. The first collections contain patches from prepared design units using the ~107K PDB structures in the Guide to Pharmacology and other uncategorized MMDS target families; with biological unit expansion and alternate conformation enumeration, it turns into over 200K prepared design units. A second collection of potential sites and alternate pockets has also been built that has just over two million patches.
OEDesignUnits must be converted to SiteHopper patches before they can be searched. The Make SiteHopper Patch Database Floe creates a searchable SiteHopper collection and accepts design units prepared using the Spruce - Protein Preparation Floe.
Locate the Make SiteHopper Patch Database Floe as follows:
Click the “Floe” button on the navigation bar to reach the Floe page.
Click on the Floes Tab.
From the Categories Floe Filters, click on the ‘Packages’ drop-down to expand the list of packages, then select the Protein Modeling Floes package.
A list of the Protein Modeling floes will now be visible to the right. Click on the Make SiteHopper Patch Database Floe.
Alternatively, you can enter Make SiteHopper Patch Database in the search bar.
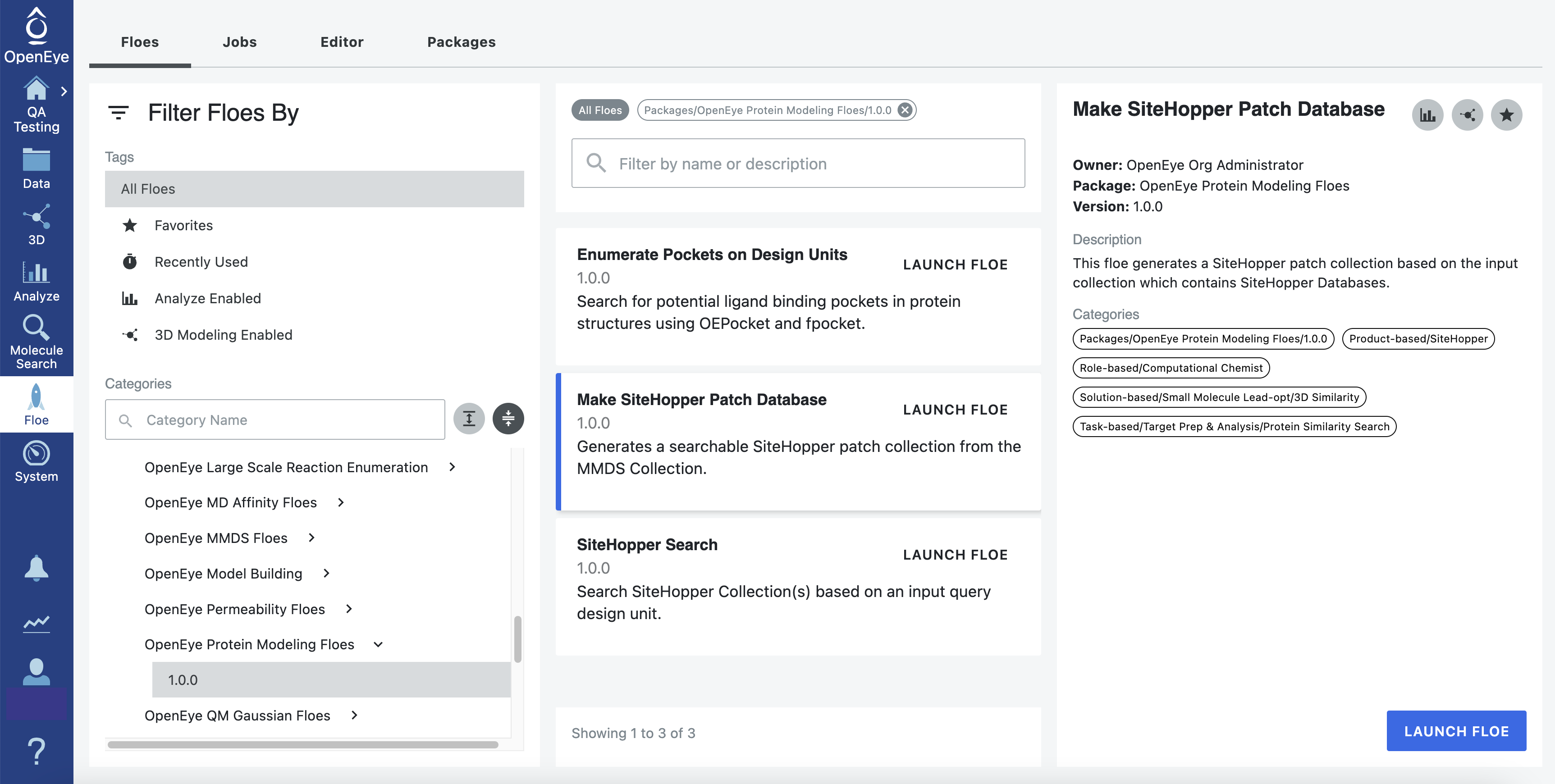
Figure 2. How to find the SiteHopper floes.
Click the “Launch Floe” button to bring up the Job Form and set the following parameters.
Inputs
Optional Dataset of Design Units: To select this collection:
Click the “Choose Input” button.
Click on Organization Data under collections in the Select Optional Dataset of Design Units pop-up window. Organization is the name of your company or organization. This name is set during Orion installation or configuration; however, an organization admin can also change the name.
Select the Spruce Prep Dataset CDK2 dataset.
Click “Use Dataset as Input.”
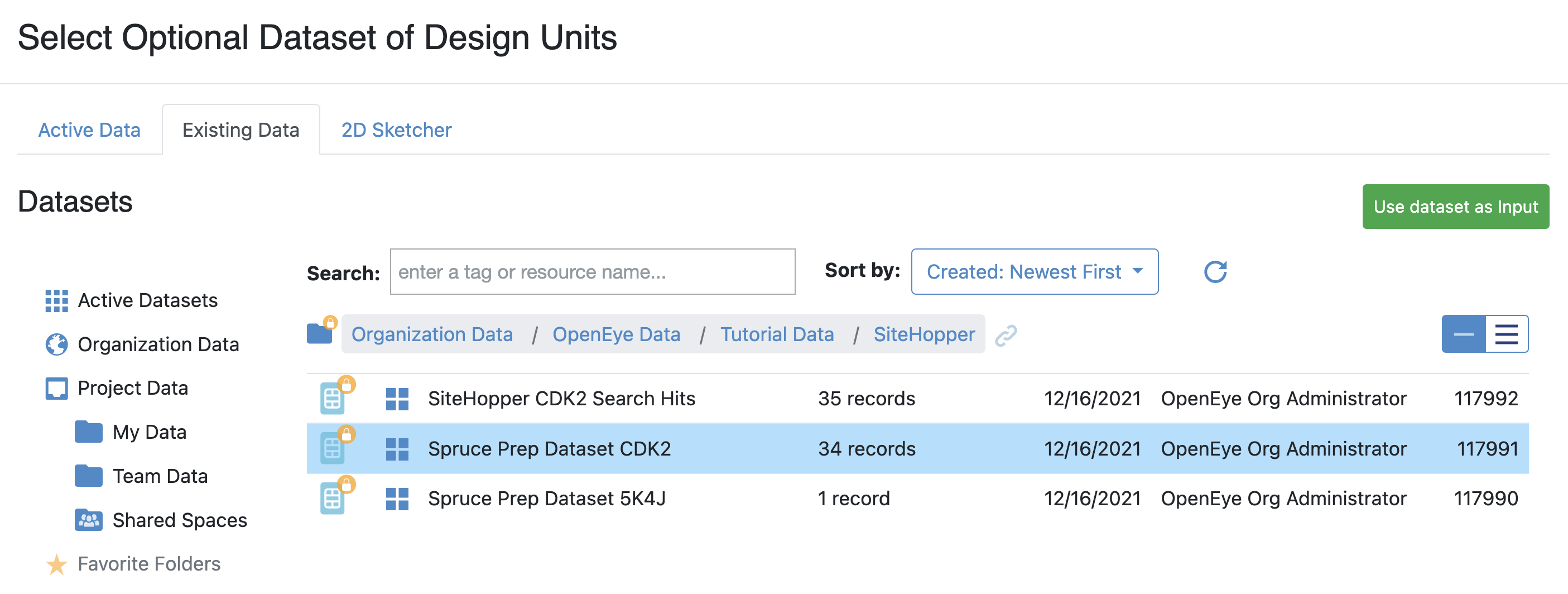
Figure 3. Select the input dataset.
Outputs
Collection Name: Enter CDK2 Patch DB Collection.
Failed Output Dataset: Enter Failed Patch Dataset.
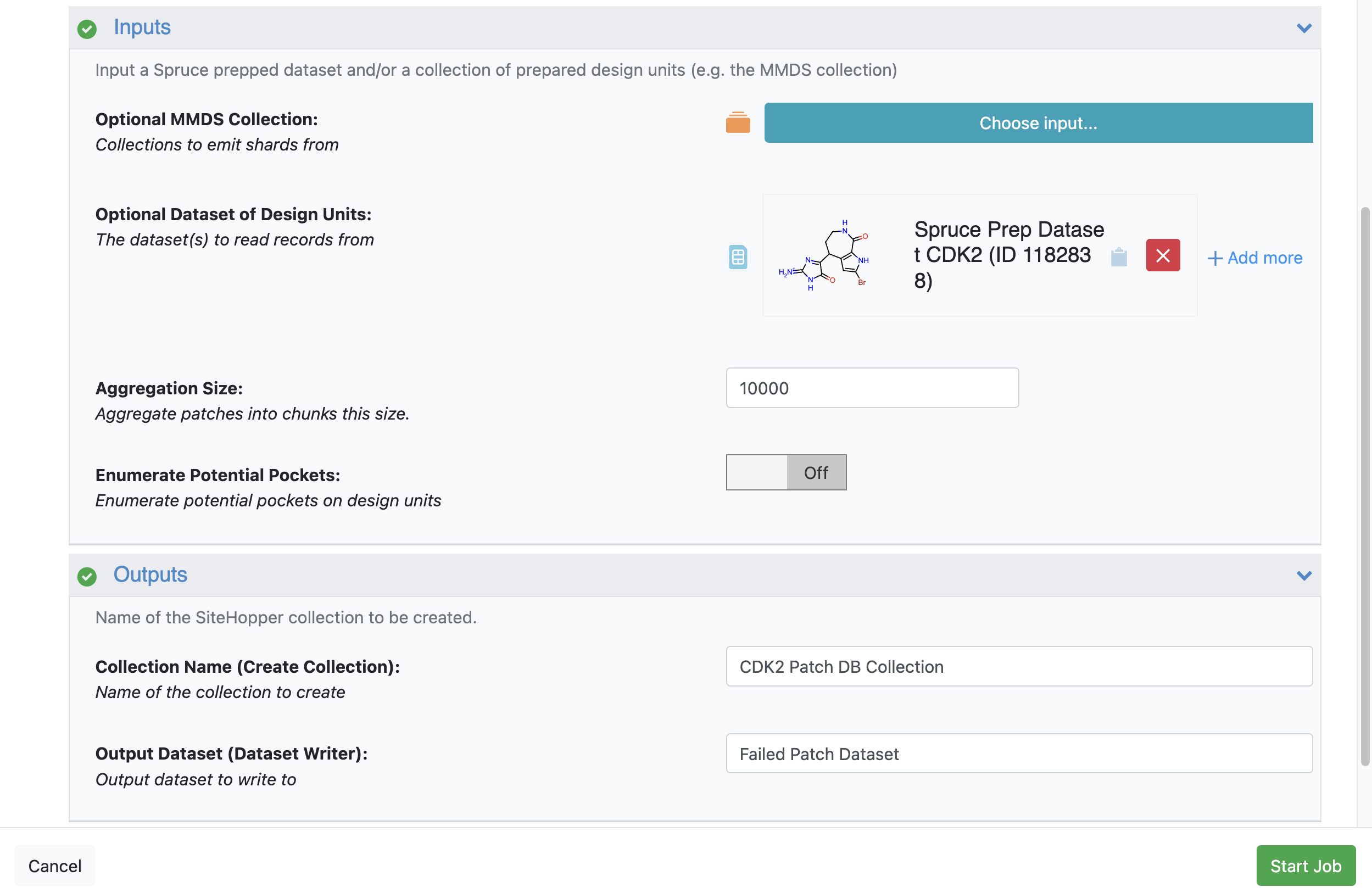
Figure 4. Job Form for the Make SiteHopper Patch Database Floe.
At the bottom of the Job Form, click “Start Job” to begin the floe. The job will take about five minutes to run. Once the floe has finished, move on to the next step of the tutorial.
Note
Switching Enumerate Potential Pockets to On creates a SiteHopper Database collection where liganded sites are skipped, but other potential binding sites are enumerated using pocket finding tools.
Prepare Query Protein
For this tutorial, use the CDK2 crystal structure 5K4J from the Protein Data Bank. To import this structure into Orion and prepare it for docking, locate the SPRUCE - Protein Preparation on the Floe page.
There is a tutorial detailing the use of that floe here: Spruce Prep Tutorial.
This dataset can also be used as a reference against a list of other PDB codes, which is how the demo dataset Spruce_prep_dataset_CDK2 was created.
View Prepared Site
Once the Spruce - Protein Preparation job finishes, make the resulting dataset active as follows:
Navigate to the Data page using the blue navigation bar.
Select Organization Data on the left-hand side of the page.
The ‘Show’ drop-down menu should have the Datasets option checked.
Click on the ‘Active Datasets’ drop-down in the Active Data Bar and deselect any active datasets.
Activate the Spruce_prep_dataset_5K4J dataset by clicking the “Dataset Activation” button to turn it from a white plus sign to a green checkmark.
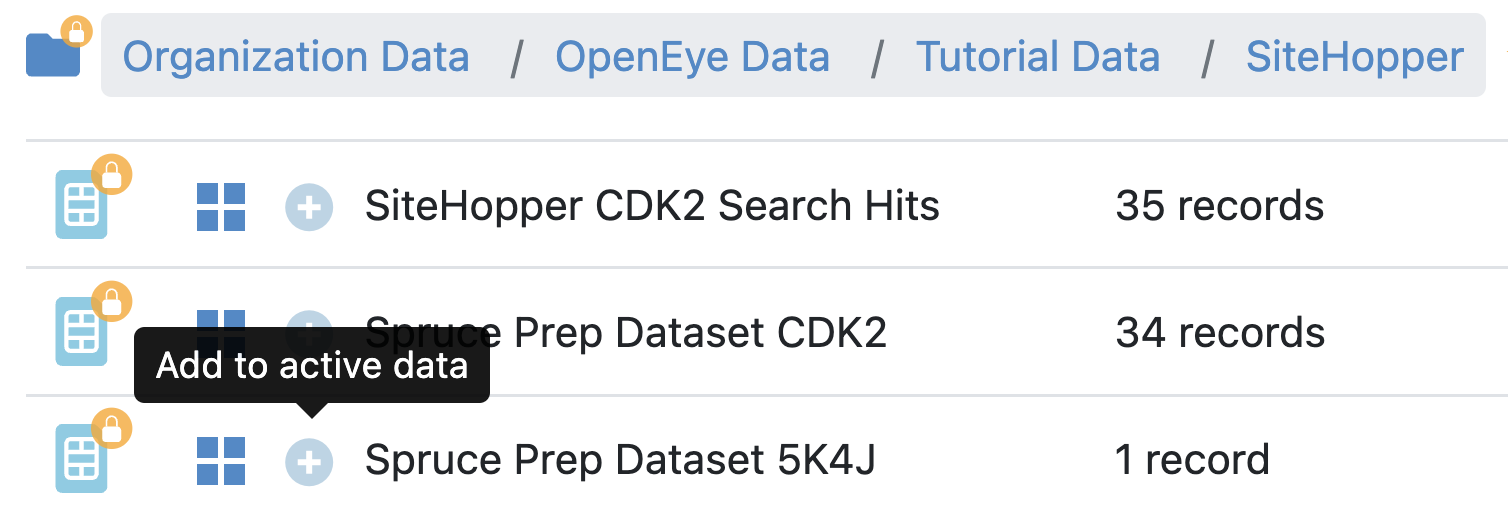
Figure 5. How to activate a dataset.
Now you can see the query in the 3D Viewer Panel.
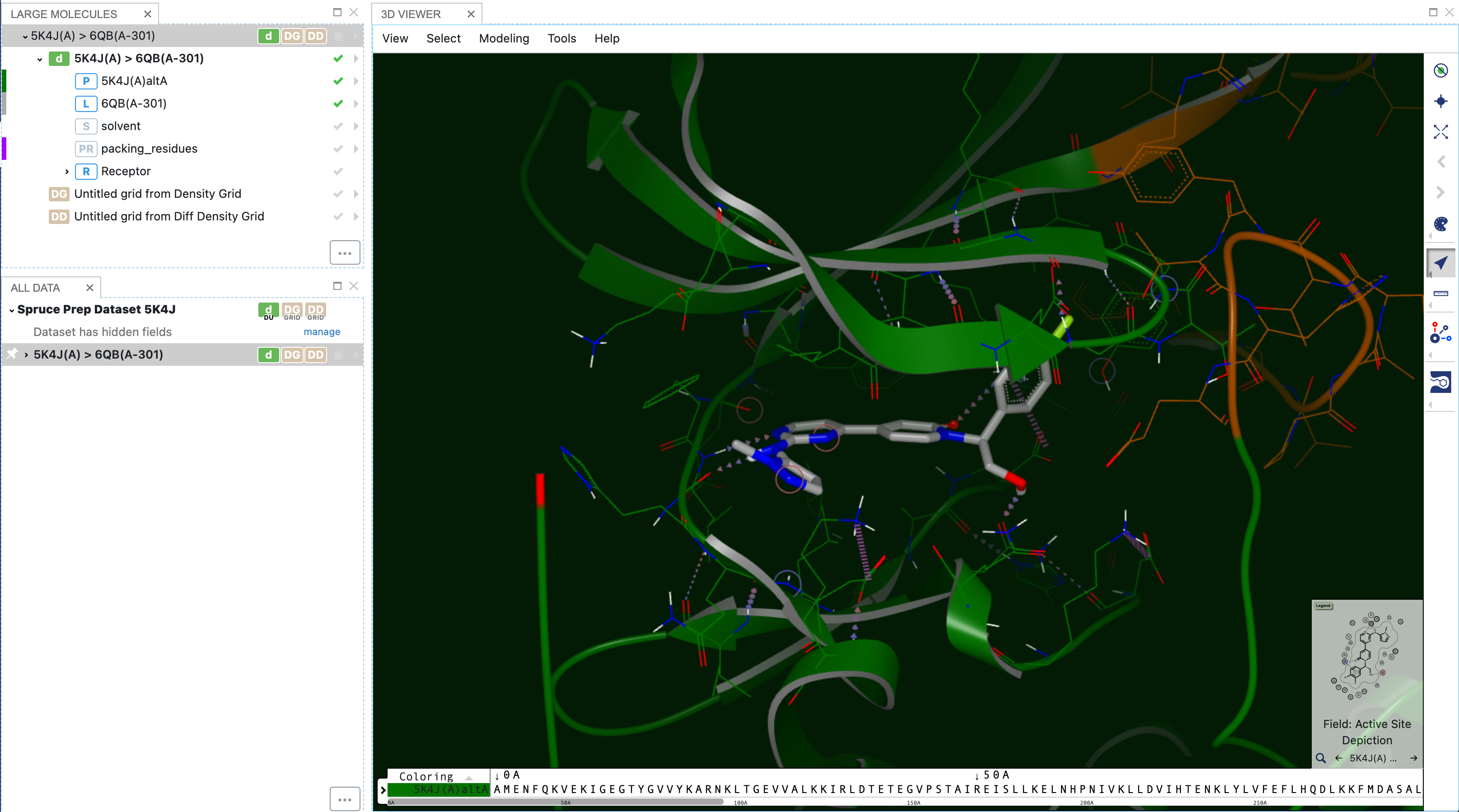
Figure 6. The query shown in the 3D Viewer window.
Search Patch Dataset
Here we will search the smaller CDK2 collection generated above; there are also MMDS prepared patch databases available for searching in the Organization data.
Locate the SiteHopper Search Floe as described for the Make SiteHopper Patch Database Floe: on the Floes Tab of the Floe page, click on the ‘Packages’ drop-down to expand the list of packages, and select the Protein Modeling Floes package to bring up the list of Protein Modeling floes. Please see Figure 2.
Click on the SiteHopper Search Floe, then the “Launch Floe” button to bring up the Job Form and set the following parameters.
Inputs
Input Dataset: Query Dataset: Spruce_prep_dataset_5K4J
This is the dataset with the protein we prepared.
Input Collections: CDK2 Patch DB Collection
This is the dataset with all the patches we created at the beginning of this tutorial.
Outputs
Output Dataset: SiteHopper CDK2 Search Hits
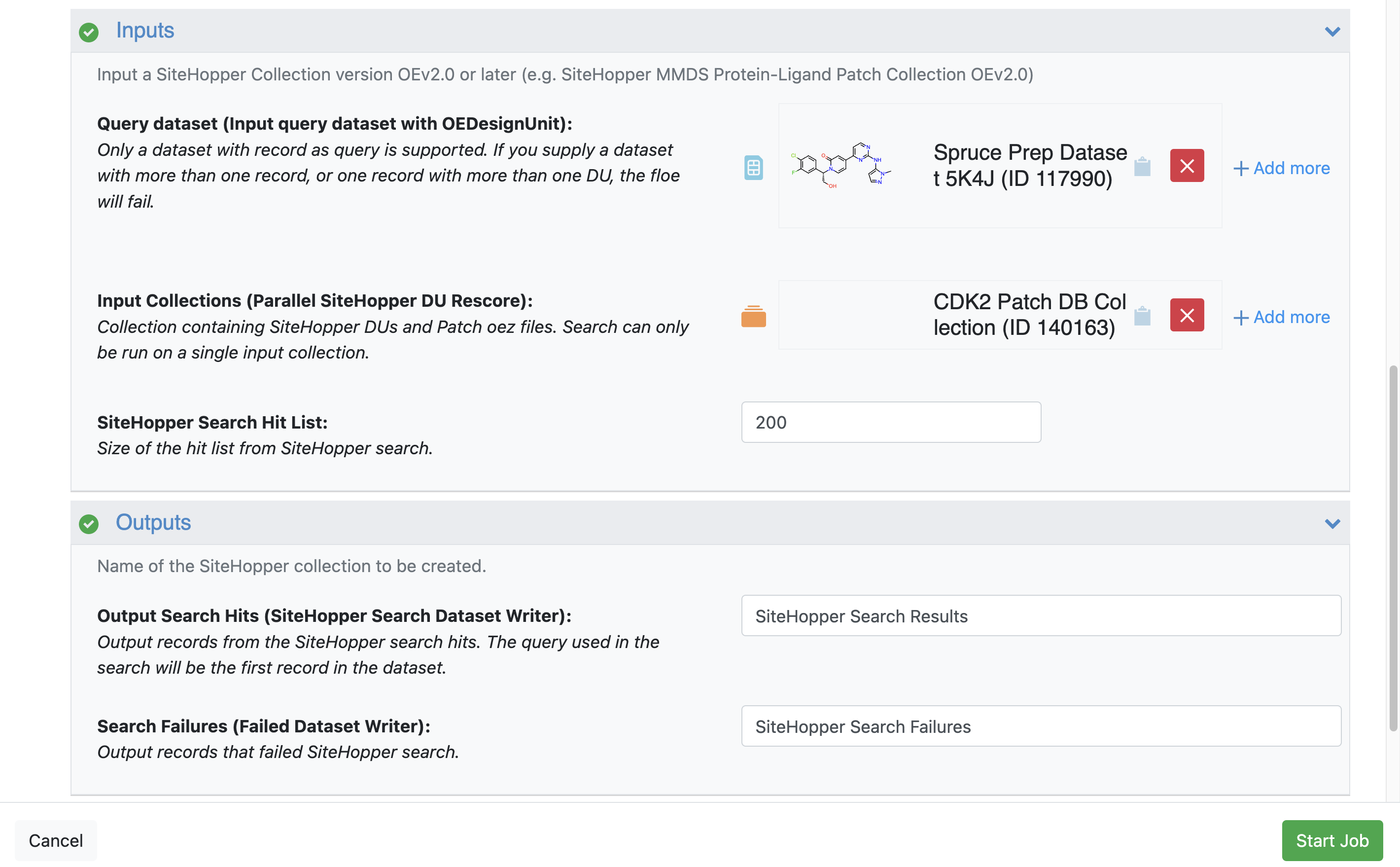
Figure 7. The Job Form for the SiteHopper Search Floe.
Once the parameters are set, click the “Start Job” button to start the floe. The job will take 5–10 minutes depending on the query, and potentially longer if the databases being searched are large.
View Results
Once the search job has completed, make the output datasets active as follows:
On the Data page, clear current active datasets by clicking on the ‘Active Datasets’ drop-down and then “Clear All.”
Activate the output datasets by clicking on the + icon (“Dataset Activation” button).
On the 3D page, the 3D results are overlaid on the input query, which is the first record in the resulting dataset. Here, one of the top results is shown with the query. This is similar to how FastROCS results are shown for small molecules.
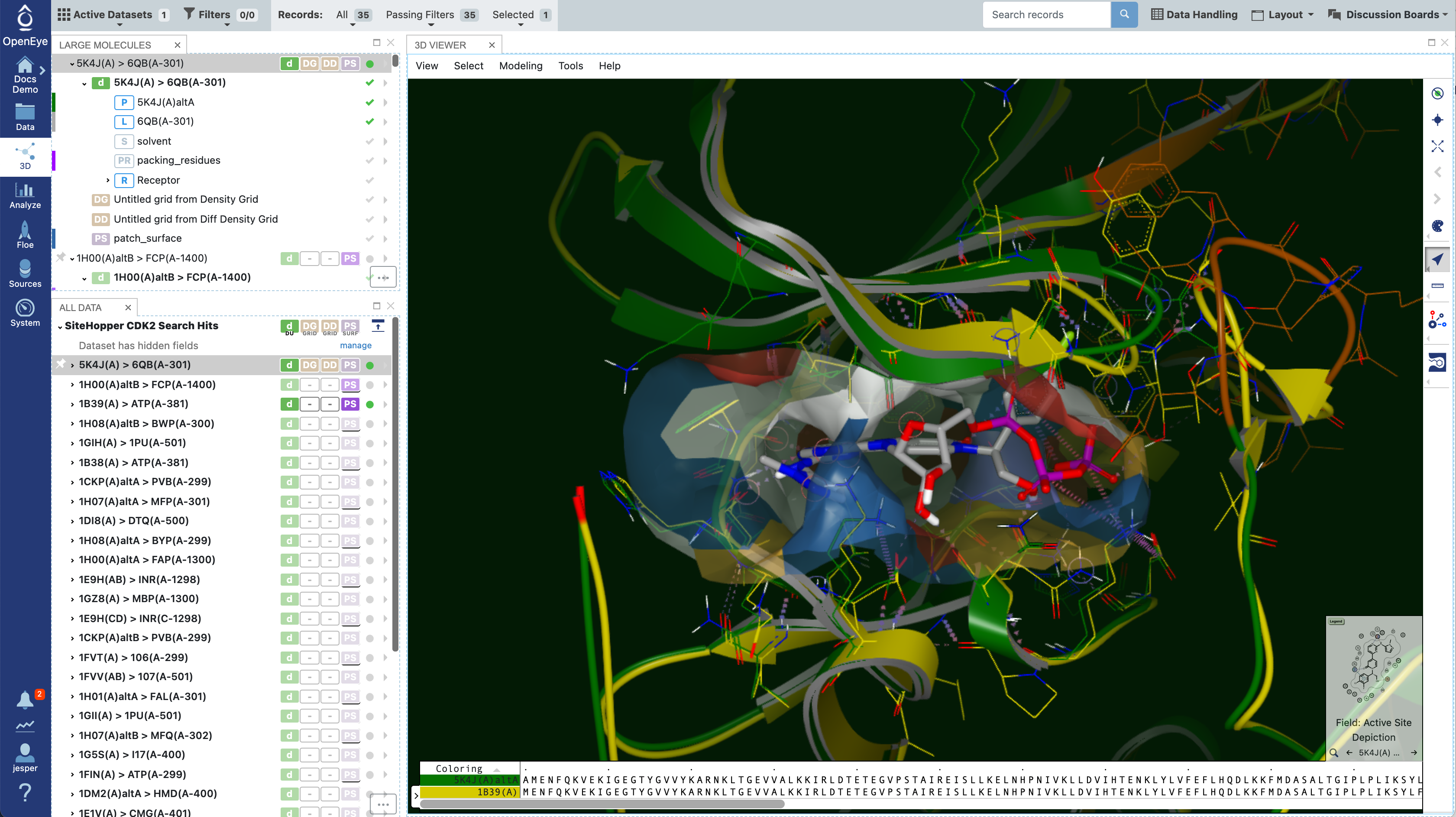
Figure 8. The query molecule overlaid with a result from the SiteHopper Search Floe.
Analysis of the scores and examination of interesting results can also be done in the Analyze page, based on SiteHopper scores and sequence similarity scores. Here, patch scores are plotted against sequence similarity. Of course, for a dataset of the same target, the sequence similarities are very high and perfect for most of the dataset. As larger databases are searched, this property becomes more interesting to explore.
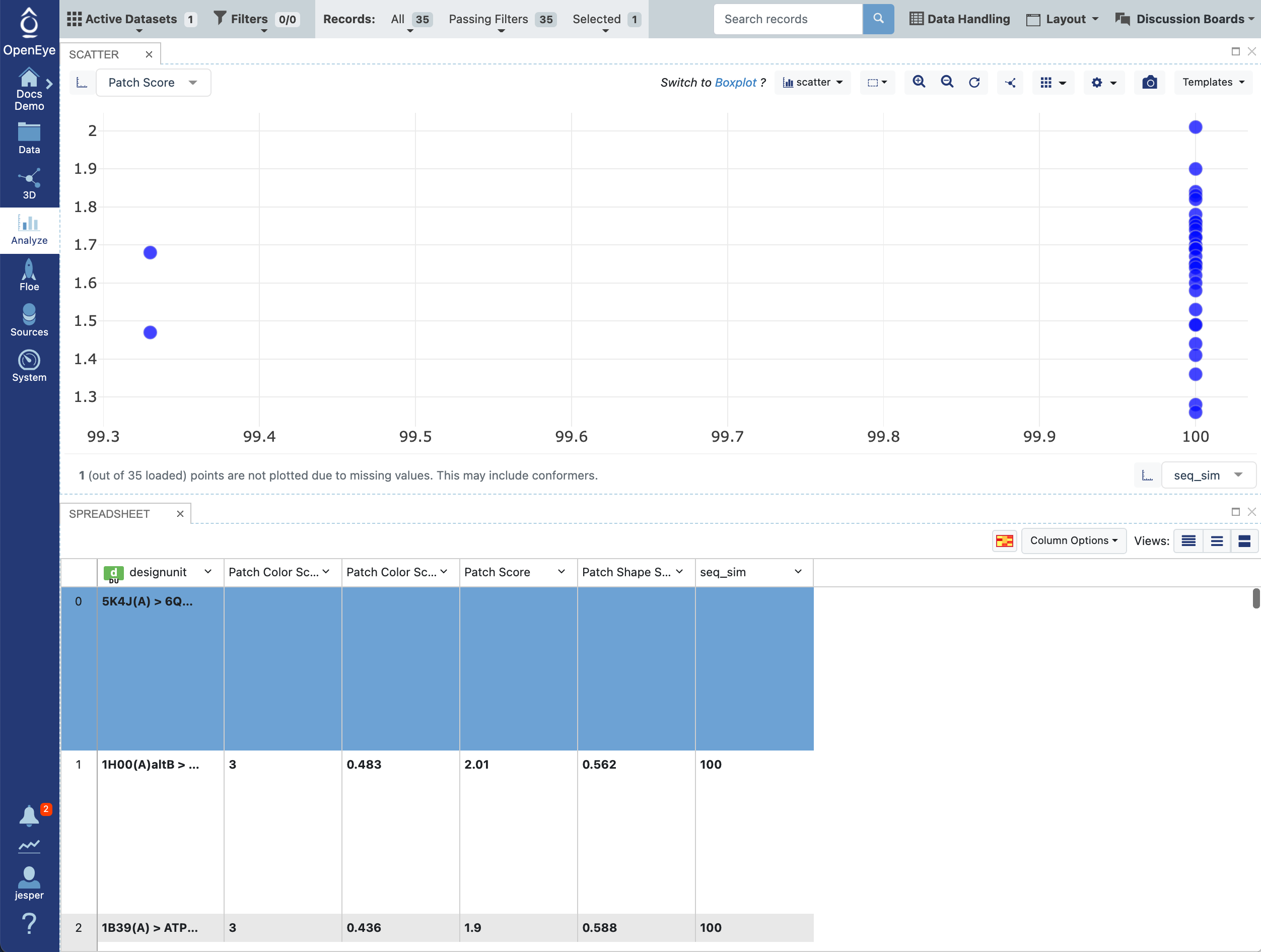
Figure 9. The Analyze page showing results from the SiteHopper Search Floe.