Solvate and Equilibrate Target Protein
Quick floe search term: CPD A1
In this tutorial, we will show you how to use the Spruce-prepared KRas protein in the MD preparation Solvate and Equilibrate Target Protein Floe.
Tip
This floe should take approximately one hour to run and cost ~$3.
Search and Run the Floe in Orion
Locate the floe in Orion
Using the blue navigation bar, go to the Floe page.
On the Floe page, click on the Floes Tab, where you will find the list of the available floes and packages.
Under the Category Floe Filters, click on the caret next to the ‘Packages’ Filter to expand the list of packages and select the OpenEye Cryptic Pocket Detection Floes package. The floes for the Cryptic Pocket Detection package will appear in the middle pane.
From this list, click on the Solvate and Equilibrate Target Protein Floe and then click the blue “Launch Floe” button in the bottom right corner to launch the Job Form as shown in Figure 1.
Provide Input Files and Parameters to Run the Floe
- Output Path:
Select the destination for your output data by specifying the Output Path.
- Input Data:
Choose the Target Protein to be the Spruce-prepared dataset for KRas.
- Output Data:
You can customize the output dataset and collection names in this parameter group.
- Force Field:
Change the force fields for the protein and ligand by selecting the desired Protein Parameters and Ligand Parameters.
- Solvent Options:
By default, this floe solvates the target protein in water-xenon mixed solvent with 150 mM xenon and 150 mM NaCl salt. By changing the Solvent Molar Fractions and Solvents input parameters, you can change the concentration of the xenon probes for a mixed-solvent simulation or set up a single-solvent simulation by changing the Solvent Molar Fractions to 1.000, Solvents to tip3p, and Salt Concentration to 150 mM.
Caution
If you change the Solvent Molar Fractions and Solvents parameters to run a single-solvent simulation, you will be able to perform only exposon analysis for cryptic pocket detection. Probe occupancy or dynamic probe binding analysis for cryptic pocket detection cannot be performed on the data generated by a single solvent simulation.
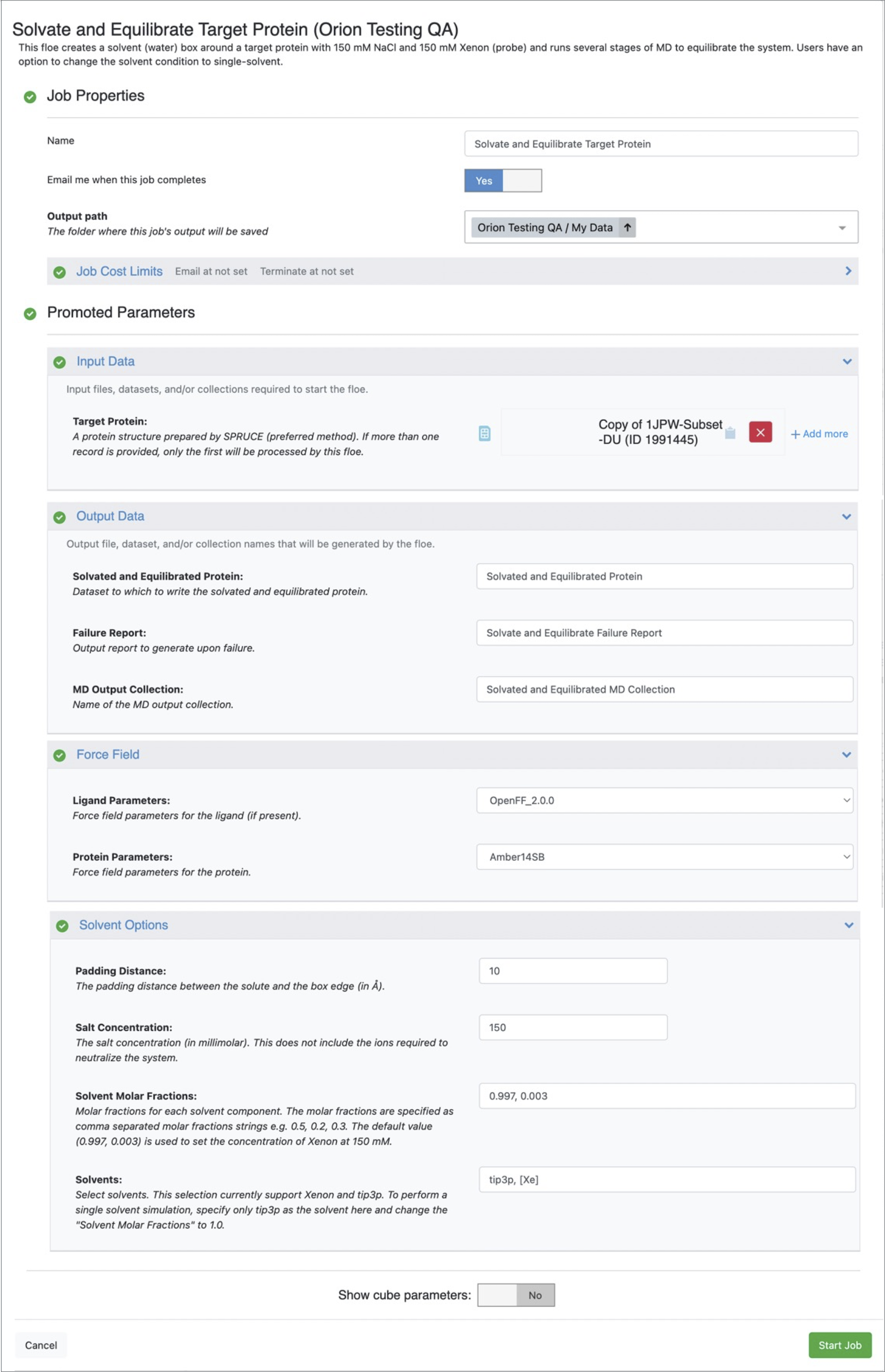
Figure 1. Job Form for the Solvate and Equilibrate Target Protein Floe.
Click on the green Start Job button.
If the job completes successfully, there should be an MD dataset, a reference PDB file containing all of the system components, an h5.tar.gz file, and a collection generated.