Release Notes, Version 2025.2.1
Release Highlights 2025.2
Small Molecule Discovery Suite Highlights
pKa Prediction Floes Package
The new pKa Prediction Floes Package predicts the dominant ionization states and pKas of small molecules. The ionization state of a drug candidate affects crucial physical properties including solubility, membrane permeability, target binding affinity, and ADMET. Therefore, using the correct ionization state for a given pH environment is very important to get accurate results from many computational methods, especially binding free energy calculations. The Generate Ionization States and Calculate LogD Floe predicts the dominant ionization state at a specified pH. The pKa Exploration Floe Floe provides a detailed report showing the titration curves and distributions of ionization states at different pH values.
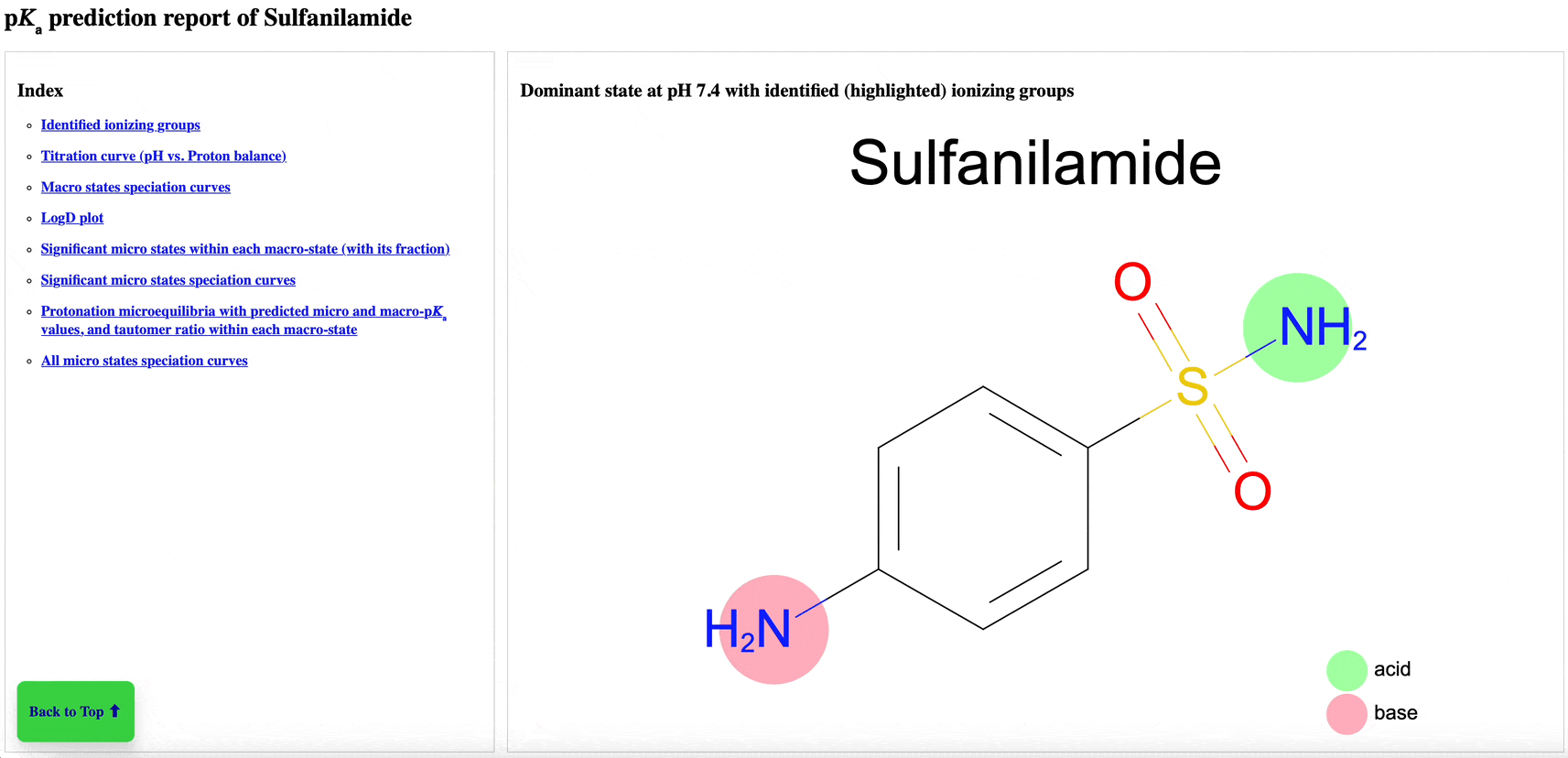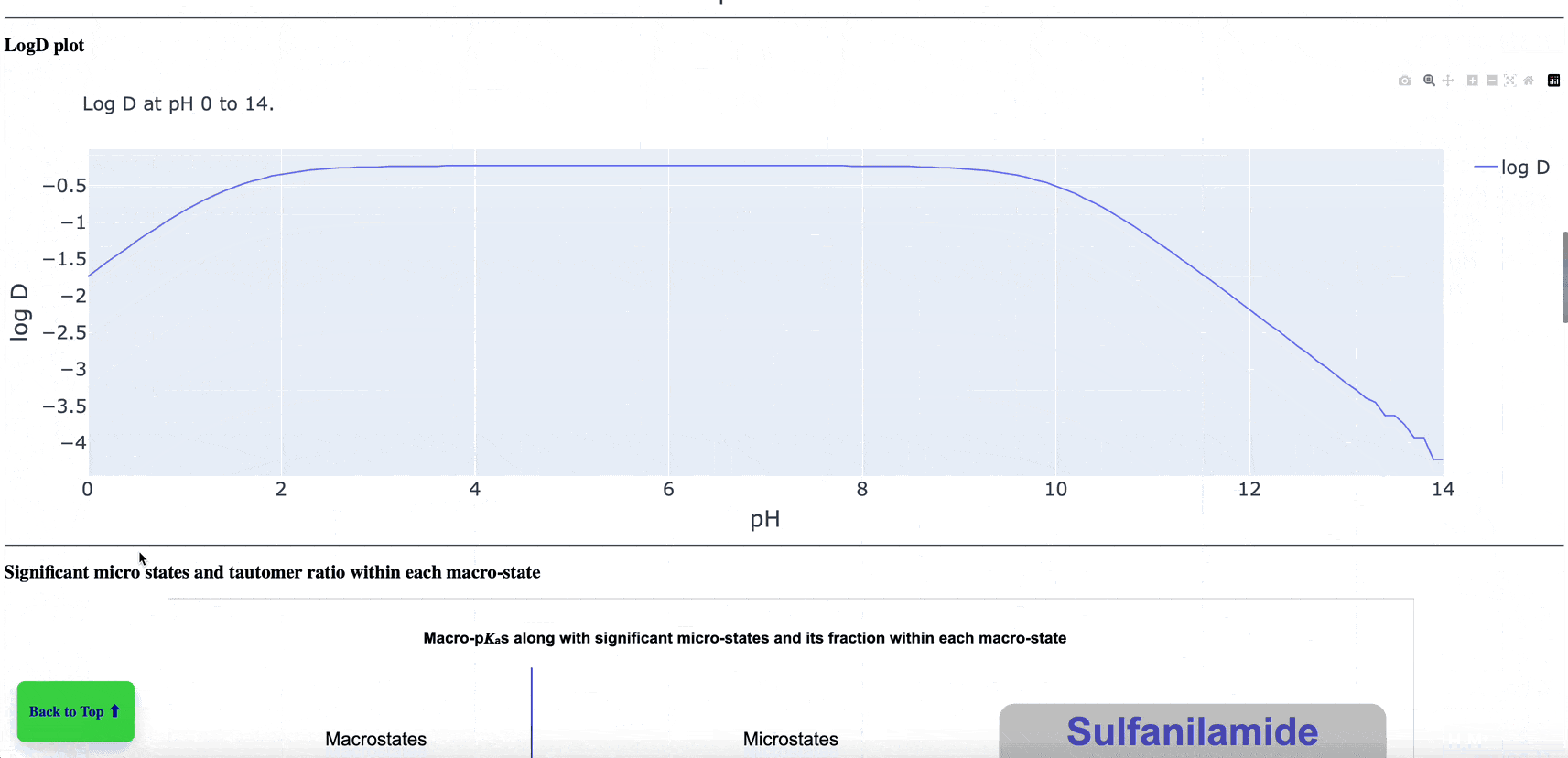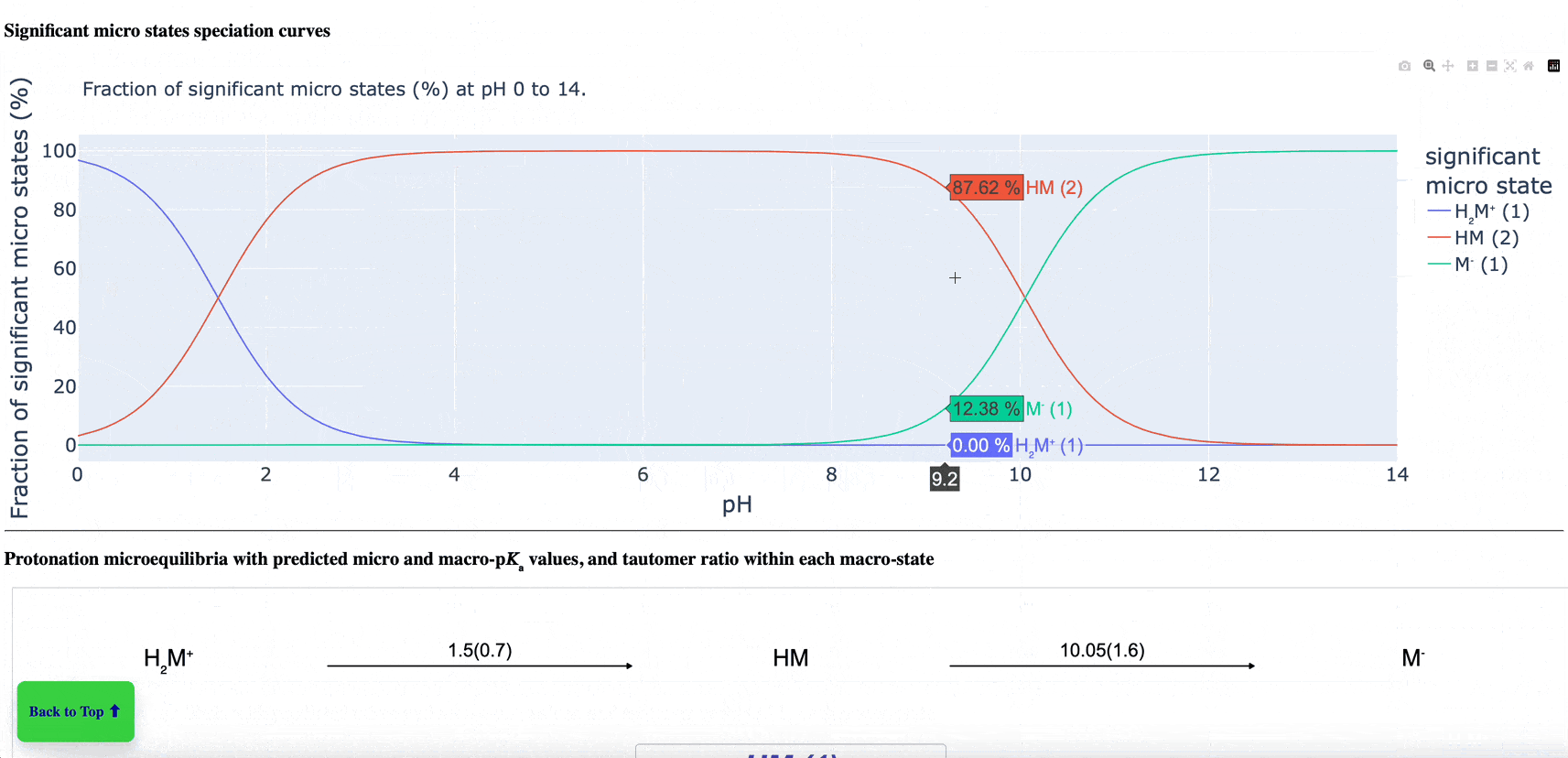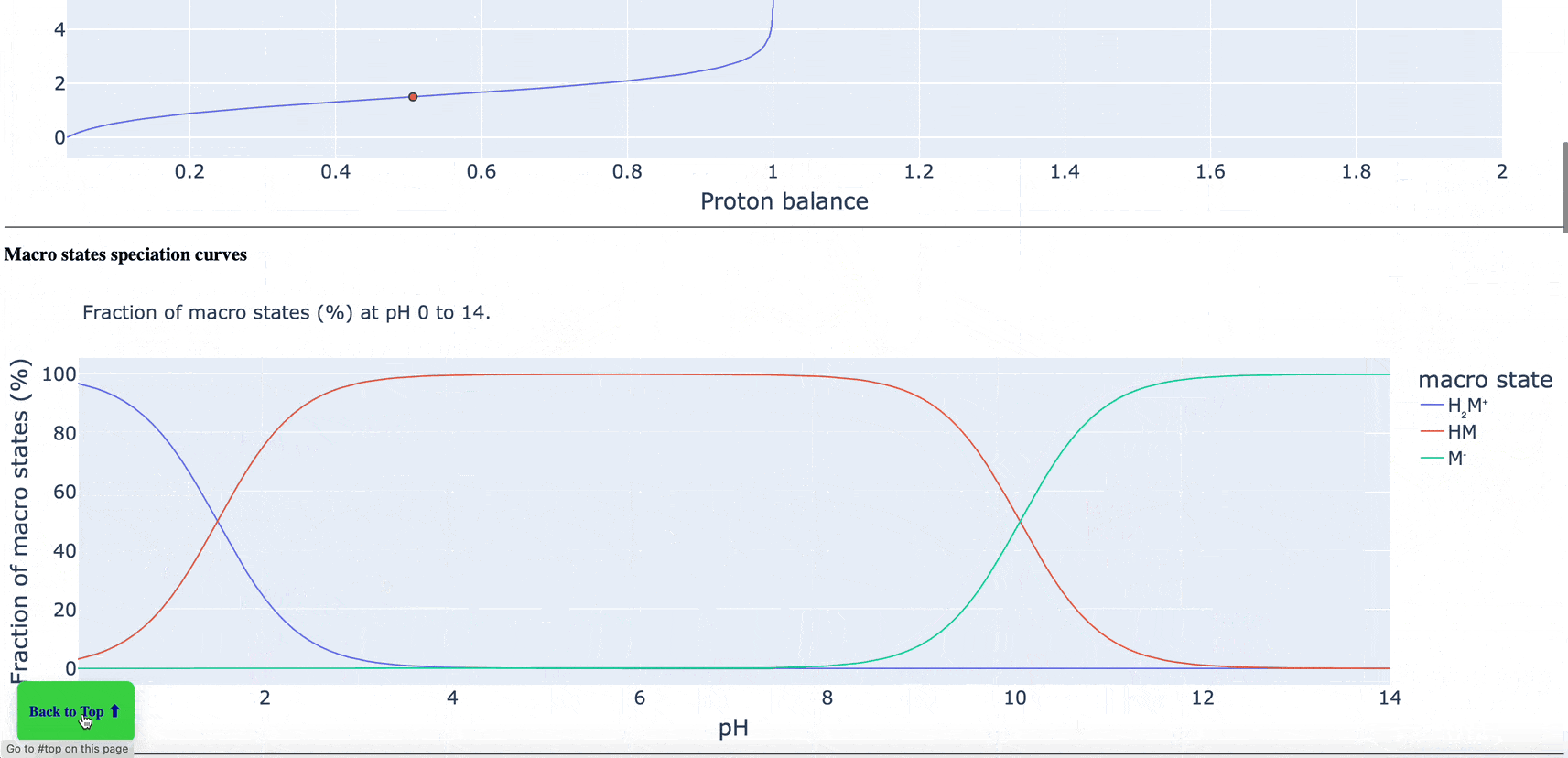
Figure 1. The floe report identifies all ionizing groups and includes the titration curve, macrostate population curves, logD versus pH, microstate population curve, and micro- and macro-pKa reactions with tautomer ratios. This animated figure shows each aspect of the report and the changes of various features in relation to others. Note: to view the animation again, please refresh your browser after the animation has finished. Individual parts of the floe report can be found in the pKa Prediction Report within the pKa Exploration Floe tutorial.
Cryptic Pocket Detection Floes Package
Ligandability Assessment for Pocket Ranking and Representative Conformation Selection
Cryptic pockets are ranked by a ligandability prediction model which is trained and validated on a curated, nonredundant dataset consisting of diverse ligand-binding pockets. The unnormalized total score and normalized ligandability score, along with the significance score, are then computed for each pocket found during cryptic pocket detection.
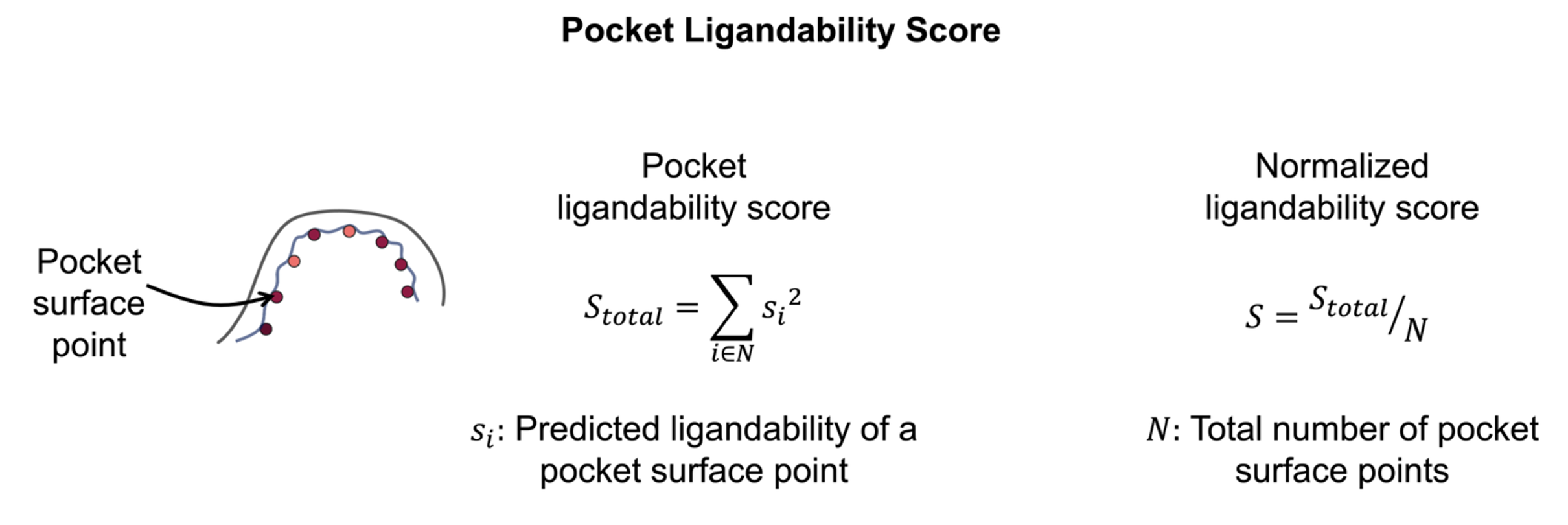
Figure 2. The pocket ligandability score prediction.
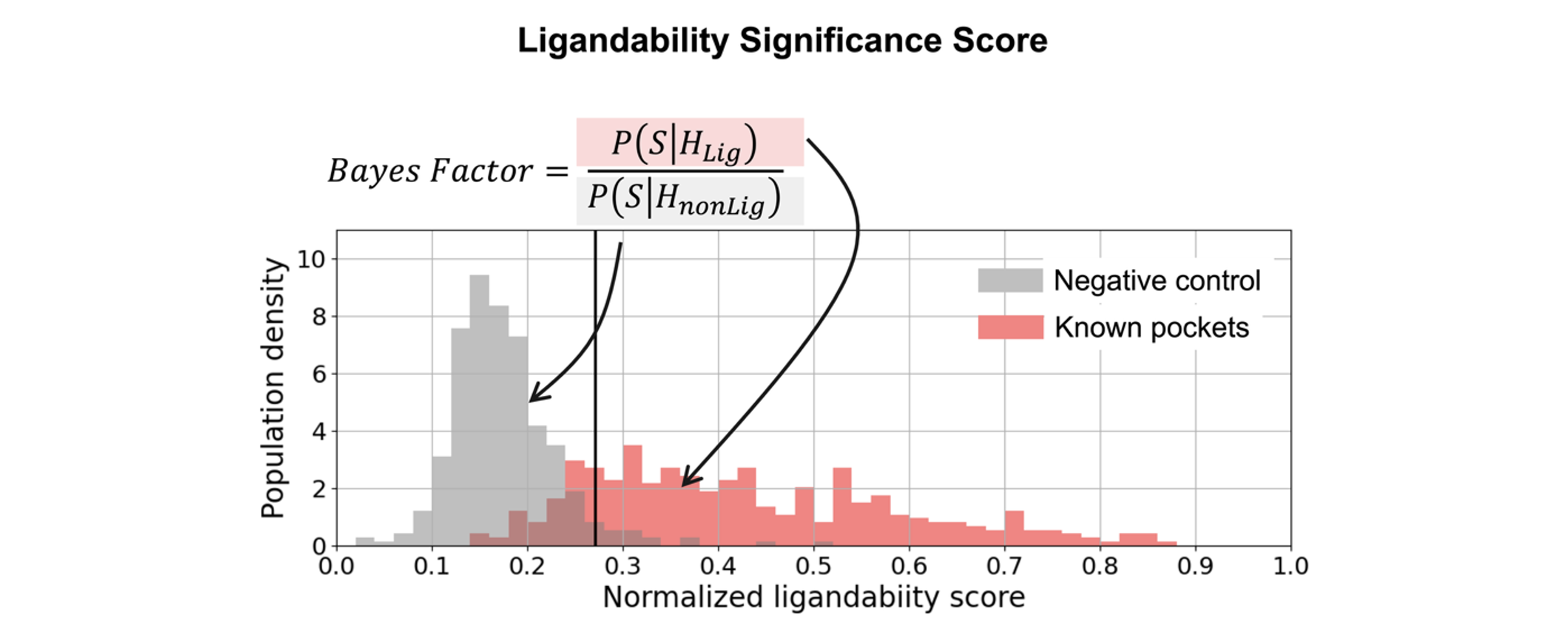
Figure 3. The assessment of significance score (Bayes factor) for the normalized pocket ligandability score.
The new Ligandable Medoids Extraction Floe extracts an ensemble of ligandable medoid conformations for a selected pocket. A binding site view can be generated for pockets detected by probe binding analysis techniques after specifying “DUM” (the component representing xenon-occupancy grids for a pocket) as a ligand in the binding generation option.
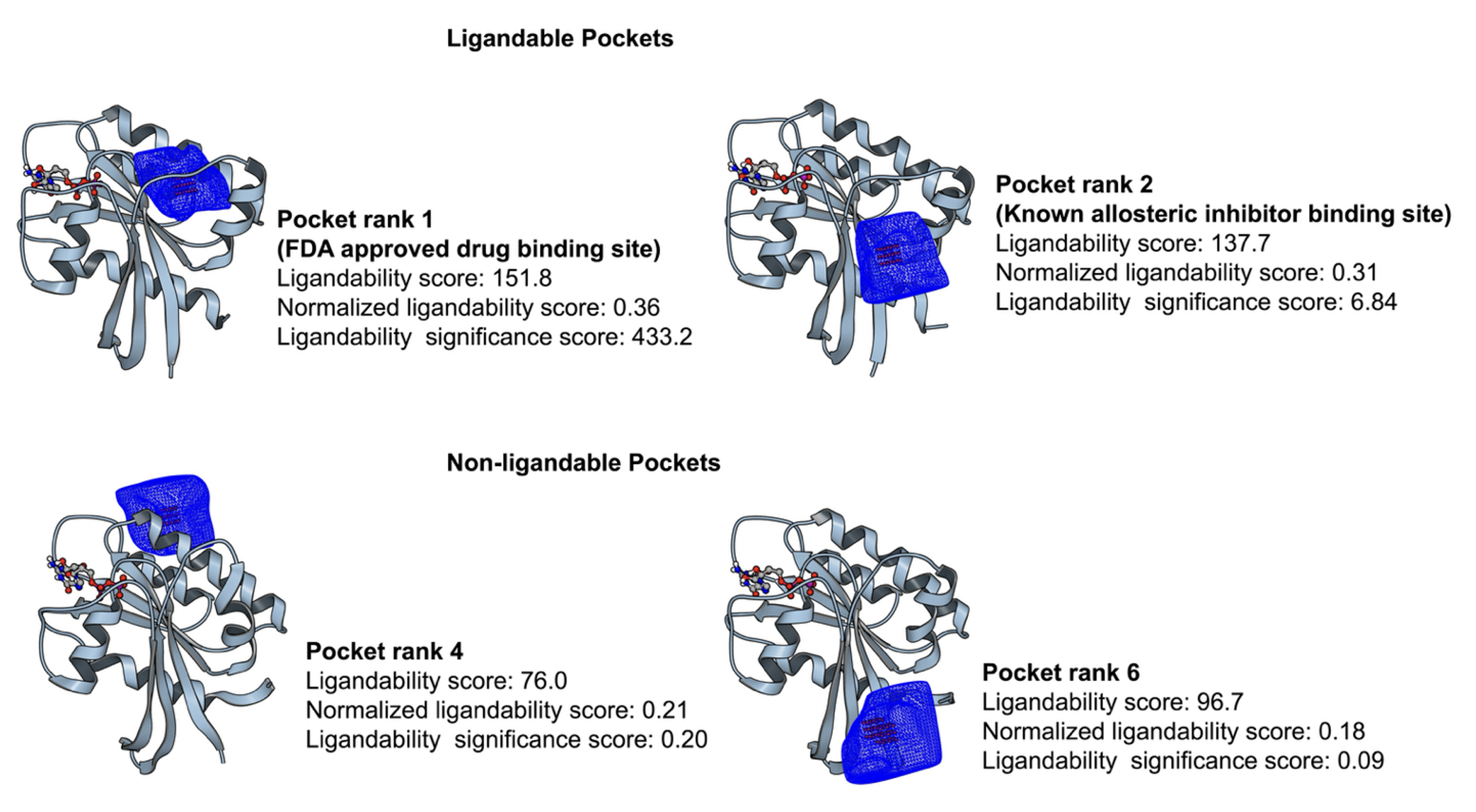
Figure 4. The ligandability score and ranking for cryptic pocket predictions.
Improved Time and Cost Efficiency for Dynamic Probe Binding Analysis
The performance of the Dynamic Probe Binding Analysis Floe has significantly improved, now being ten times faster and five times lower in cost.
Detailed Release Notes, Version 2025.2.1
3D QSAR Models
v1.2.0 August 2025
General Notice
This package is built using
OpenEye-orionplatform==6.5.1,OpenEye-toolkits==2025.1.0, andOpenEye-Snowball==0.30.0.
Floe Updates
The floe now allows users to select a set of 3D models to build.
Two new 3D models, ROCS-GPR-NO-2D and EON-GPR-NO-2D, have been added. Unlike ROCS-GPR and EON-GPR, these two GPR models only use 3D Tanimoto as descriptors.
The floe now accepts kcal/mol and kJ/mol as potency units.
The default cross-validation method has been switched from “leave one out” to “random” to accommodate for a larger training set.
The option Include 2D in COMBO has been added to allow the user to decide whether to incorporate 2D-GPR into final model predictions.
A new parameter section, Cube Memory Parameters, has been added to allow users to increase the memory of some cubes for relatively large training sets to avoid cube memory errors.
Grids corresponding to color atom probes have been added to the output dataset, along with contour values used for generating high and low surfaces.
The domain grid has been deleted from the output dataset.
The Generate ROCS Query parameter has been removed. ROCS query output will be generated by default along with ROCS-kPLS model interpretation.
A new parameter, Selected Models, has been added, allowing for the selection of kPLS models to interpret.
A new parameter, Percentage for Surface Contour Values, has been added to give the user more flexibility to choose contour values for high and low surfaces.
A new parameter section, Cube Memory Parameters, has been added to allow users to increase the memory of some cubes for relatively large training sets to avoid cube memory errors.
A new parameter section, Cube Memory Parameters, has been added to allow users to increase the memory of some cubes for relatively large training sets to avoid cube memory errors.
A new parameter section, Cube Memory Parameters, has been added to allow users to increase the memory of some cubes for relatively large training sets to avoid cube memory errors.
Classic Lead Optimization Release Notes
v0.16.2 August 2025
General Notice
This package is built using
OpenEye-orionplatform==6.4.1,OpenEye-toolkits==2025.1.1, andOpenEye-Snowball==0.30.1.
Floe Updates
The default memory for the single-receptor Posit Cube has been increased to 4*1.8*1024 in the POSIT - Ligand Guided Small Molecule Posing Floe.
v0.15.1 February 2025
General Notice
This package is built using
OpenEye-orionplatform==6.2.0,OpenEye-toolkits==2024.2.0, andOpenEye-Snowball==0.29.1.
Floe Updates
The CHOMP - Generate BROOD Fragment Database Floe has been improved as follows to improve usability and robustness:
A Report Only parameter has been added that would produce a quick report of fragments without attempting to generate the database.
The memory requirement per fragment has been reduced by keeping track of just a single source molecule instead of five.
Instead of failing due to lack of required memory, the floe now attempts to complete the task with the amount of fragments that it could process.
An Input Existing Brood Database parameter has been added to enable passing in an existing database, fragments from which should not be included in the newly created database.
The default setting has been adjusted for fragments larger than nine heavy atoms to be included in the database if the fragment is contained in at least two of the source molecules.
The zipped database, in .TAR.GZ format, is saved by default.
The floe no longer fails to write fragments when input molecules are missing titles.
The BROOD - 3D Fragment Replacement Floe has gone through the following changes:
The Floe Report now provides comprehensive information on the hit list generation along with the details of the clustering and the hit molecules.
The default memory for the hit list generation cube has been increased to 48*1.8*1024
The FreeForm - Bound Ligand Configurational Free Energies with SZYBKI Floe now properly uses the bound ligand from the primary molecule field when an input ensemble is provided by the user through a separate field.
The BROOD - DB File to Collection Floe now generates a report if it fails to complete successfully for some reason.
The BROOD - Query Builder Floe now generates a report if it fails to complete successfully for some reason.
The POSIT - Ligand Guided Small Molecule Posing Floe now properly respects user choice for the Relaxation Mode parameter. The default for the Relaxation Mode has also been changed to Clashed, to ensure that generated poses are clash free by default.
The Calculate Atropisomerism Floe now assigns AM1BCCELF10 charges by default and provides an option to change the default behavior.
Legacy Release Notes
v0.3.1 August 2025
General Notice
This package is built using
OpenEye-toolkits==2025.1.0,OpenEye-orionplatform==6.4.1,OpenEye-Snowball==0.30.0, andOpenEye-orionmdcore==2.5.4.
Minor Changes
The Automated Cryptic Pocket Detection with Probe Occupancy Analysis, Combined Probe Binding Site Analysis, Probe Occupancy Analysis, Dynamic Probe Binding Analysis, and Exposon Analysis Floes will finish with the job status “Success” if no cryptic pockets are detected for a given target instead of a “Failed” completion. A Failure Report will be generated listing the potential reasons and next recommendations when no pockets are detected for a target.
If users provide optional Important Residues input while running the Automated Cryptic Pocket Detection with Probe Occupancy Analysis, Combined Probe Binding Site Analysis, Probe Occupancy Analysis, Dynamic Probe Binding Analysis, and Exposon Analysis Floes, the center-of-mass distance between the user-defined Important Residues and Pocket Residues will be reported for each pocket in the Pocket Receptors output dataset generated by these floes.
Major Changes
The Filter Trajectory Features Data Cube (previously named Preprocess Trajectory Analysis Data) has a seven-to-tenfold lower memory requirement. The Curate Cryptic Pockets Cube (previously named Cryptic Pockets Reporter Cube) has more than a twofold lower memory requirement. Collectively, these changes reduce the chances of failure due to memory error associated with cryptic pocket floe report generation while running the Automated Cryptic Pocket Detection with Probe Occupancy Analysis, Combined Probe Binding Site Analysis, Probe Occupancy Analysis, Dynamic Probe Binding Analysis, and Exposon Analysis Floes.
The Dynamic Probe Binding Analysis is more than twelvefold faster in this version. The run cost for this floe is now expected to be fivefold less than the previous version.
A new floe, Combined Probe Binding Site Analysis, that combines Probe Occupancy Analysis and Dynamic Probe Binding Analysis has been introduced. Running this floe eliminates the need to separately run the Probe Occupancy Analysis and Dynamic Probe Binding Analysis Floes for cryptic pocket detection.
A new OEligandability-score based pocket ranking has been introduced. For each pocket reported by the Automated Cryptic Pocket Detection with Probe Occupancy Analysis, Combined Probe Binding Site Analysis, Probe Occupancy Analysis, Dynamic Probe Binding Analysis, and Exposon Analysis Floes, the total ligandabilty score; the ligandability score normalized by total surface points as well as by receptor volume; and the ligandability significance score will be reported in the Pocket Receptors output dataset.
v0.2.7 March 2025
General Notice
This package is built using
OEToolkits 2024.1.1,Orion Platform 6.1.2, andSnowball 0.28.0,Orion Molecular Dynamics (MD) Core 2.4.2.
Minor Changes
The Cryptic Pockets Reporter Cube has a significantly lower memory requirement. This reduces the chances of failure due to memory error associated with cryptic pocket floe report generation while running the Automated Cryptic Pocket Detection with Probe Occupancy Analysis, Probe Occupancy Analysis, Dynamic Probe Binding Analysis, and Exposon Analysis Floes.
Temporary checkpoint files and collection are deleted after the cryptic pocket analysis floes finish. This includes an auto-cleanup for the Automated Cryptic Pocket Detection with Probe Occupancy Analysis, Probe Occupancy Analysis, Dynamic Probe Binding Analysis, and Exposon Analysis Floes.
v0.2.5 September 2024
General Notice
This package is built using
OEToolkits 2024.1.1,Orion Platform 6.1.2, andSnowball 0.28.0,Orion Molecular Dynamics (MD) Core 2.4.2.
Minor Changes
A fix has been made to allow analysis of protein structures with protonated histidine residue names (HIE, HID and HIP).
An issue with display of pocket residues in the cryptic pocket analysis floe report has been fixed. It now displays the pocket residues when the protein chain IDs do not begin with “A” or when the protein chain IDs do not follow the alphabetical order.
v0.2.4 July 2024
General Notice
This package is built using
OEToolkits 2024.1.1,Orion Platform 6.1.2, andSnowball 0.28.0,Orion Molecular Dynamics (MD) Core 2.4.2.
Minor Changes
A correction has been made in the axis of the 2D free energy projection on the normal modes.
A default value for mutual information cut-off for pocket detection using the Dynamic Probe Binding Analysis and Exposon Analysis Floes is set to 0.00. This parameter is also now exposed to users under Cube Parameters.
Faster receptor creation in the Probe Occupancy Analysis, Dynamic Probe Binding Analysis, and Exposon Analysis Floes makes them more time efficient.
The documentation now includes an FAQ section.
v0.2.2 April 2024
General Notice
This package is built using OEToolkits 2023.2.3, Orion Platform 6.1.2, and Snowball 0.27.1.
Note
There is a known incompatibility between datasets created by previous versions of Cryptic Pocket Detection and datasets used by this version. If you need assistance with this issue, please contact Support@eyesopen.com.
Major Changes
The Automated Cryptic Pocket Detection with Probe Occupancy Analysis Floe is introduced for automated, end-to-end workflows for cryptic pocket detection with probe occupancy analysis using an input design unit prepared using Spruce.
Three advanced Floes (Probe Occupancy Analysis, Dynamic Probe Binding Analysis, and Exposon Analysis) automate the cryptic pocket analysis process by integrating analysis of weighted ensemble protein sampling data, iterative clustering, cryptic pocket detection, and receptor creation.
Weighted ensemble protein sampling data storage has been significantly refactored, which reduces the overall size by half and supports faster data find and read.
Two new options have been added, Maximum Number of Selected Modes and Mode Filtering Property, in the Calculate Normal Modes Floe that allow for automatic rank and select modes based on specific mode properties. This feature is turned On by default in the Automated Cryptic Pocket Detection with Probe Occupancy Analysis Floe.
Minor Changes
In addition to the Automated Cryptic Pocket Detection with Probe Occupancy Analysis Floe, individual sub-floes starting from the Solvate and Equilibrate Target Protein Floe to three cryptic pocket analysis Floes (Probe Occupancy Analysis, Dynamic Probe Binding Analysis, and Exposon Analysis) can be sequentially run for protein sampling and cryptic pocket analysis.
Now the Run a Normal Mode-Guided Weighted Ensemble MD Simulation Floe generates and stores data in a V2 ShardCollection.
The outputs have been changed from the Run a Normal Mode-Guided Weighted Ensemble MD Simulation Floe. Now the output dataset contains multiple records (one record per iteration) and only contains summary statistics for the simulation. All simulation data (e.g., weighted ensemble data, MD trajectories, simulation files) are stored in the output shard collection. The collection is now self-contained and required as input for downstream analysis Floes.
The Subsystem Selection String option has been substituted with Environment Selection String in the Calculate Normal Modes Floe for ease of use. Now the system-environment framework is enabled by default in the Floe.
The Continue a Normal Mode-Guided Weighted Ensemble MD Simulation Floe now takes the collection generated by the Run a Normal Mode-Guided Weighted Ensemble MD Simulation Floe as the input to resume the weighted ensemble protein sampling simulation.
v0.1.2 September 2023
General Notice
This package is built using OEToolkits 2023.1.0, Orion Platform 5.1.1, and Snowball 0.26.0.
Minor Changes
Fixes an issue related to “select string” parameter format in the Calculate Normal Modes floe.
Enables support for non-canonical amino acids in the Solvate and Equilibrate Target Protein floe.
A minor typographical correction in documentation for the Calculate Normal Modes floe.
v0.1.0 July 2023
General Notice
This is the first release of OpenEye Cryptic Pocket Detection Floes.
This package is built using OEToolkits 2023.1.0, Orion Platform 5.1.0, and Snowball 0.26.0.
Generative Design Floes
New Tutorial August 2025
A new tutorial was added to the documentation: Tutorial: Library Enumeration for Hit Identification.
Software features are the same as with v2.4.1 (January 2025).
Legacy Release Notes
v6.6.0 August 2025
General Notice
This package was built using
OpenEye-orionplatform==6.6.0andOpenEye-toolkits==2024.1.1.
Functional Changes
The records_per_shard parameter on the ParallelRecordsToCollectionCube cube has been promoted in the Dataset to Collection Export ETL floe.
v6.3.2 January 2025
V6.3.2 is a rerelease of 6.3.1, with no change in content.
v6.3.1 September 2024
A bug has been addressed in which SMILES data in an input file following CXSMILES data caused a parsing error.
A bug has been addressed when exporting datasets to CSV files. All columns, or a subset thereof, can now be exported in cases when previously only Molecule and Molecule.Titled would have been exported.
v6.3.0 August 2024
A bug has been addressed in which SMILES data in an input file following CXSMILES data caused a parsing error.
v6.2.0 June 2024
Support for ShapeQuery (.sq) files has been added.
Support for CXSMILES data has been added.
Created collections have been defaulted to v1.
A bug has been addressed by which some english language words (such as “BOTH”) could be interpreted as molecules.
v6.1.2 February 2024
Columns can now be optionally ignored when exporting datasets.
v6.0.0 September 2023
The version number has been synced with openeye-orionplatform.
The Dataset to File Export ETL floes now preserve the Molecule Title when exporting to CSV files.
v2.3.0 July 2023
This version increases the minimum versions of the requirements:
OpenEye-orionplatform==5.1.0openeye-toolkits>=2022.2.2
v2.1.3 February 2023
A floe for uploading large or small files from a URL has been added
v2.1.2 November 2022
The orion-platform requirement has been updated to 4.5.2
v2.1.1 September 2022
The orion-platform requirement has been updated to 4.4.1
An issue where
orionclient.types.Shard.download()could receive less data than expected has been fixed.An
IOErrorwill be raised andorionclient.types.Shard.download_to_file()will catch and retry.
v2.1.0 July 2022
An issue with importing convert_path has been fixed.
The etl-floe classifications has been updated to use the new
PurePosixPathclassificationsThe
brieffield has been added and thedescriptionfield has been made more descriptive on all ETL floes.Requirement changes:
The Sphinx requirement has been updated to
3.5.4and moved to requirements.txt.The jinja2 requirement has been updated to
3.0.3.The orion-platform requirement has been updated to
4.4.0.The openeye-toolkits requirement has been updated to
2022.1.1.The openeye-floe-pkg-tools requirement minimum version has been updated to
1.0.3.
The etl-floe classifications has been updated to use the new
PurePosixPathclassifications.The “brief” field was added and the “description” field was made more descriptive on all ETL floes.
v2.0.2 February 2022
Updated orion-platform to 4.3.0.
Converting datasets containing image data to csv now results in that field containing “<Image>”.
v2.0.1 December 2021
The openeye-toolkits requirement has been updated to 2021.2.0
v2.0.0 November 2021
The Collection Resize floe has been removed
v1.2.9 November 2021
The orion-platform requirement has been updated to 4.2.5
v1.2.8 October 2021
The orion-platform requirement has been updated to 4.2.4
A bug where the metadata of a collection was not being preserved when running the Collection Resize ETL floe has been fixed.
A required parameter collection_type in the ParallelCollectionResizeCube of the Collection Resize ETL floe has been promoted.
v1.2.7 October 2021
orion-platform has been updated to 4.2.0
A failure caused by csv files containing no data for some columns has been fixed.
IUPAC names are no longer parsed, resulting in fewer column splits and ETL speedup.
A failure parsing lists of numbers has been fixed.
A bug in the naming of split columns has been fixed.
A failure when converting a record to OEB has been fixed. Fields with data that cannot be properly attached as generic data to a molecule are now skipped by default.
v1.2.6 June 2021
The orion-platform requirement has been updated to 4.1.0 (mono-package)
The OpenEye-toolkits requirement has been updated to 2021.1.1
A regression in perceiving basic molecular properties for SMILES and CSV has been fixed.
v1.2.5 June 2021
The orion-platform requirement has been updated to 4.0.0 (mono-package)
Orion Platform >= 4.0.0 will not work with stacks older than 2021.1 due to usage of new APIs
The OpenEye-toolkits requirement has been updated to 2020.2.4
A new Converter,
SmilesFileConverter, has been added.When converting CSV and SMILES format files to records, the original smiles are stored on output records.
ETL floe documentation has been improved
v1.2.4 November 2020
The orion-platform requirement has been updated to 3.1.0
BaseDatasetFieldAddOrReplaceCubehas been added.If no file extension is provided to
DatasetWriterCube’s output file parameter, the file format is nowOEDB.OpenEye-floe==0.12.0
Adds support for versioning of WorkFloes.
Adds support for ordering of Parameters in the Orion UI.
An issue that resulted in Serial Cube Group initializer ports not incrementing the counts has been resolved.
OpenEye-drconvert==1.1.2
Bump OpenEye Toolkits
OpenEye-orionclient==3.1.0
Bump OpenEye Toolkits
OpenEye-datarecord==3.0.1
Bump OpenEye Toolkits
v1.2.3 August 2020
orion-platform has been updated to 3.0.0
An issue with too frequent flushing in
DatasetUpdaterCubehas been fixed.The
DatasetAppenderCubenow finalizes datasets.OpenEye-datarecord==3.0.0
The implementation of datarecord has mostly moved to C++ in the toolkits.
A field type
DesignUnitfor storing OEDesignUnits has been added.
OpenEye-drconvert==1.1.0
Support for OEDesignUnit has been added
OpenEye-orionclient==3.0.0
Added OEDesignUnit support for dataset upload and download
Support for uploading OEDU files to datarecords has been added to
upload()orionclient.types.Datarecord.recordsnow yieldsOEMolRecord
v1.2.2 April 2020
orion-platform has been updated to 2.4.5
OpenEye-orionclient==2.6.3
The efficiency and reliability of
ShardCollection.list_shardshas been improved.
v1.2.1 March 2020
orion-platform has been updated to 2.4.4
A bug in
RecordsToCollectionCubehas been fixed.Improvement made to shard retries.
v1.2.0 February 2020
The base OS image has been upgraded to Amazon Linux 2
The environment has been upgraded to Python 3.8.1
Package documentation has been added
orion-platform has been updated to 2.4.2
Improvements since 2.1.0 include:
Support for the
OEZformat has been added to the following cubes:CollectionResizeCubeCollectionToRecordsCubeRecordsToCollectionCube
orionplatform.cubes.files.RecordsToRecordFileConverternow uploads its files in parts reducing the disk usageNew parameters, which optionally enable metric collection for cubes have been added.
Sets lower bound on OpenEye-toolkits>=2019.10.2
A duplicate of
output_tagsin theDatasetWriterCubeOptions parameter group has been removed.OpenEye-Drconvert==0.7.2
Support added for Float and Int vectors
Drconvert CLI now supports going from records to mols
v1.1.1 October 2019
orion-platform 2.1.0
Improves reliability of uploading shards and writing shards of records
Adds
limitparameter toShardReaderCubeto allow streaming only some shards from a collection.
v1.1.0 August 2019
orion-platform 2.0.0
Improves reliability of collection floes
Updated version of drconvert (0.6.20) correctly handles conformer data
v1.0.0 July 2019
orion-platform 1.0.2
v0.1.29 April 2019
orion-platform 0.1.16
Collection resize floe
v0.1.28 February 2019
Bugfixes
drconvert 0.6.16
No longer sets
OEIsomericConfTeston file formats that are not mol2 or sdf
v0.1.27 February 2019
Add Dataset Copy floe
v0.1.26 February 2019
Orion Platform 0.1.14
openeye-drconvert>=0.6.15,<0.70
Performance improvements
Bugfixes
Fixes
ArchiveConverterCube
v0.1.25 January 2019
Orion Platform 0.1.13
openeye-drconvert>=0.6.13,<0.70
Calls
flush()before returning records fromMolFileConverter
openeye-datarecord>=0.12.7,<0.13.0
Adds
flush()toOERecordto write all cached values to bytes.
openeye-schemagen>=0.3.3,<0.4.0
Minor cleanup
v0.1.24 December 2018
orion-platform 0.1.12
Summary of changes relevant to ETL floes:
openeye-drconvert>=0.6.12,<0.7.0
Handle whitespace in SD data
openeye-orionclient>=0.6.10,<0.7.0
Makes Orion Client more robust to handling API changes
v0.1.23 November 2018
orion-platform 0.1.11
v0.1.22 November 2018
orion-platform 0.1.10
Summary of changes relevant to ETL floes:
openeye-schemagen>=0.3.2,<0.4.0
survive SD data consisting of only whitespace
openeye-datarecord>=0.12.5,<0.13.0
add more testing for metadata; Serialize/deserialize round trips. ORION-6328
add Meta.Flags.Predicted metadata to datarecord. Minor cleanups. ORION-6328
v0.1.21 October 2018
orion-platform 0.1.9
Summary of changes relevant to ETL floes:
openeye-schemagen>=0.3.1,<0.4.0
delimiters back to ordering by descending priority
openeye-orionclient>=0.6.9,<0.7.0
0.6.9
Adds headers to improve debugging capabilities
0.6.8
Adds orionclient.exceptions.OrionTimeout exception that indicates that
a request couldn’t be made to the specified Orion within the expected time
openeye-floe>=0.6.15,<0.7.0
0.6.15
Fixes issue where parallel workers would incorrectly propagate failures when run in Orion
v0.1.20 October 2018
add package uuid
v0.1.19 October 2018
orion-platform 0.1.8 * fix memory leaks when reading dataset * improve robustness of collection handling
v0.1.18 September 2018
orion-platform 0.1.7 orion-client fixes: * avoid writing too many records at once * fix issue retrieving records when sizes of records are not consistent
v0.1.17 September 2018
orion-platform 0.1.6
v0.1.16 September 2018
sync job system tags with backend/frontend
bump orion-platform to 0.1.5 to force min versions
remove default tag that appears to not be used
Added continuation char BUGZID: ORION-6089
Allowing non-SDData formats for etl export BUGZID: ORION-6089
v0.1.15 September 2018
Needed to rev version because of Orion failing inspection due conflict with a previous failed upload of this version:
“Workfloe Record Collection to Dataset with version:’0.1.14’ and uuid:’934d77a2-c1a1-421f-8daf-5b8ca6325fbf’ already exists for user with ID 1”
v0.1.14 September 2018
Add package setup, tasks, tests, and release notes
invoke upload-packageputs package in S3 for devops to load onto stacksUpdate orion-platform requirement to 0.1.4, which pulls in toolkit, drconvert and related packages
Add record collection floes
pKa Prediction Floes
v0.1.0 August 2025
General Notice
This package is built using
OpenEye-orionplatform==6.5.1,OpenEye-toolkits==2025.1.0, andOpenEye-Snowball==0.30.0.This is the first release of OpenEye Small Molecule pKa Prediction Floes package.
Protein Modeling Release Notes
v1.0.0 February 2025
General Notice
This is a new package as a part of the Orion Floes reorganization. It includes Floes previously found in the Biomodeler Floes package.
This package is built using
OpenEye-orionplatform==6.3.1,OpenEye-toolkits==2024.2.1, andOpenEye-Snowball==0.29.1.The SiteHopper Search Floe has improvements to the GPU prescreen to generate hits with higher accuracy. Options have been added to filter hits based on a sequence similarity cutoff and a patch score cutoff.
Small Molecule Modeling Release Notes
v2.1.1 August 2025
General Notice
This package is built using
OpenEye-orionplatform==6.5.1,OpenEye-toolkits==2025.1.0, andOpenEye-Snowball==0.30.0.
Floe Updates
The OEDocking - Dock into an Active Site for Virtual Screening Floe has the following updates:
There is an option to fail optimized poses for excessive bending of aromatic rings in the Refined and STMD quality options.
The AUTO docking mode now uses FRED or HYBRID for docking based on the similarity of the molecule to the bound ligand.
FRED is used if the receptor is apo, or if the molecule has low similarity to the bound ligand.
HYBRID is used if the receptor is holo, and if the molecule has high similarity to the bound ligand.
You can now use the same protein mask in the Docking, Optimization, and Check Clashes Cubes.
These changes have been made in the STMD mode. You can now:
Generate 50 poses using exhaustive docking with FRED.
Perform pose clustering before optimization (in addition to one after optimization) using the ChemGauss4 Score for ranking the pose before optimization, and an RMS threshold of 1.0 for deduplication.
Perform deduplication of a pose as it is, without doing any overlay.
The SZYBKI - Ligand Minimization in a Rigid Active Site Floe and SZYBKI - Ligand Minimization in a Flexible Active Site Floe have a new parameter, bent_ring_check, to set whether or not to fail the optimization when there is excessive deviation from planarity in aromatic rings.
v2.0.1 February 2025
General Notice
This package is built using
OpenEye-orionplatform==6.2.0,OpenEye-toolkits==2024.2.0, andOpenEye-Snowball==0.29.1.
Floe Updates
The OEDocking - Dock into an Active Site for Virtual Screening Floe has been improved to provide clash-free docked poses by default. A new parameter quality has been added to the floe regarding the quality of output desired. The default setting Refined refers to optimizing the generated docked poses if there were any existing clashes. The Classic option does not perform any post optimization for clashes. The STMD option produces multiple clash-free poses, as recommended for Short Trajectory Molecular Dynamics.
The SZYBKI - Small Molecule Minimization Floe has a new parameter, H-only Optimization, enabling optimization of only the hydrogen atoms in the molecule.
The SZYBKI - Single point MMPBSA Floe now contains a new parameter, Use input charges, enabling the floe to work with existing charges in the molecule and bypassing assignment of AM1BCCELF10 charges.
The Dataset Classification – Bemis-Murcko Floe includes a bug fix for auto-detecting field names, which previously caused the floe to hang.
The EON - Shape and Electrostatic Similarity Alignment Floe has been modified to use cubes from snowball. The following snowball cubes are used, replacing the local cubes:
Legacy Release Notes
v0.30.2 Sept 2025
Cube Updates
A fix was added to the Input PDB Codes for Spruce Cube to prevent failure due to endpoint changes at the RCSB
v0.30.1 August 2025
General Notice
This package is built using
OpenEye-orionplatform=6.6.0,OpenEye-toolkits==2025.1.0,OpenEye-szmap==1.7.1.1, andOpenEye-szybki==2.8.0.1.
New Cubes
The new Docking Method Selector (Single Receptor) Cube has been added to select the docking method (FRED or Hybrid) based on similarity to the bound ligand and to emit the record to the corresponding port.
Cube Updates
OMEGA cubes now have the option to use Thompson sampling.
The Omega Conformation Cube now has an option to turn on Thompson sampling for OMEGA torsion driving.
A new OMEGA mode, ROCS-TS, generates conformers suitable for ROCS calculations using Thompson sampling.
The Generate 3D Cube now uses OEConformerBuilder, a Thompson sampling based method, to generate 3D structures for input molecules.
The Slice Ensemble Cube has the option to perform slice ensembles with or without RMS overlay.
The following cubes all use a protein mask from the receptor by default:
The following cubes have the option of whether or not to fail optimization for excessive bending of aromatic rings:
The Check clashes (Individual) Cube and Check clashes (Series) Cube have the option to choose the types of clashes to ignore: None, Hydrogen, or Any.
The Check Optimized Pose Cube emits a clashed status of each pose. It also emits a failure log if all poses are clashing or if they have deviated too far after optimization. By default, a hydrogen clash is allowed by this cube.
The Fixed-Protein Ligand Minimization Individual Cube and Fixed-Protein Ligand Minimization Series Cube emit the RMSD of optimized poses.
The Fixed-Protein Ligand Minimization Series Cube only sends the record to the failure port if none of the conformers of the molecule could be optimized.
The Brood DB Reader Cube can now handle tarballs created on macOS containing AppleDouble files (with the prefix ‘._’).
The Exhaustive Docking (Single Receptor) Cube has a new option to not fail when an apo receptor is passed for
HYBRIDmode, and switch to useFREDmode instead.
v0.29.1 February 2025
General Notice
This package is built using the following dependencies:
openeye-orionplatform>=6.2.0,<7.0.0.OpenEye-Toolkits>=2024.2.0.OpenEye-szmap>=1.7.1,<2.0.0.OpenEye-szybki>=2.8.0,<3.0.0.
New Cubes
New Check clashes (Individual) and Check clashes (Series) Cubes have been added that enable checking if there are clashes between a ligand and the target protein.
A new Check Optimized Pose Cube has been added that enables checking if a ligand pose has moved too far out due to optimization.
A new Dock Output Quality preference Cube has been added that enables selection of the quality desired in docking output.
New EON Overlay, EON Overlay (Multi Query), and Combine EON Queries Cubes have been added that enable electrostatic overlays using the new Eon toolkit.
Cube Updates
The following issues have been fixed in the Generate Chomp 2D Fragments Cube:
Input molecules without a title are given a title of Molecule.
The cube now properly uses the text contents from the file coming through the smarts_port.
The following features have been added to the Build 2D BROOD Database Cube:
A new Report Only parameter produces a quick report of fragments without attempting to emit the database fragments.
A new init port accepts existing database collections, fragments of which are ignored in the newly built library.
A new info port emits information about fragments accumulated during library building.
Behavior of the following cubes related to CHOMP has been improved to track memory usage and process the maximum amount it can handle instead of failing due to lack of memory.
The Chomp Report Cube has been improved to provide more comprehensive information.
The Brood Hit list Cube now emits comprehensive information on hit list building through a summary port.
The Brood Report Cube now provides comprehensive information on hit list generation, along with the details of the clustering and the hit molecules.
The Sage force field in the following cubes now points to the latest supported Sage version (2.2.1) in the associated toolkit:
The constant for conversion from KT to kcal has been updated from 0.59 to 0.592 in both the MMPBSA (Series) and MMPBSA (Individual) Cubes.
The Freeform Conf Prepare Ensemble Cube now properly uses the bound conformer when an input ensemble of given molecules is passed through a field other than the primary molecule field.
The following cubes now accept molecules with up to 40 heavy atoms as query molecules:
The Spruce DU to Mol Cube now fails if it does not find the specified components.
The Loop Builder Cube now adds a conformer ID for each generated conformer.
The Spruce Minimize Design Unit Cube Cube now recalculates interactions after minimization.
v0.28.0 July 2024
General Notice
This package is built using the following dependencies:
openeye-orionplatform>=5.1.1,<7.0.0.OpenEye-Toolkits>=2024.1.1.OpenEye-szmap>=1.7.0,<2.0.0.OpenEye-szybki>=2.7.1,<3.0.0.
Updated to latest version of FPocket:
fpocket==4.2
New Cubes
The following new Cubes have been added to support functionality of the new OpenEye Bioisostere TK.
A new Tautomer Enumeration Cube has been added that provides the number of reasonable tautomers of a molecule.
Cube Updates
Redundancy in the available
Charge typeshas been removed in the Assign Partial Charges Cube.Ligand force fieldtypes built in to the following Cubes have been simplified for usability.The Maximum Number of Conformers parameter in the following Cubes now takes a value of 0, corresponding to generating all possible conformers.
A new parameter has been added to the following Cubes to set a nonbonded interactions cutoff.
The Enhanced Stereo Info parameter in the Enumerate Stereo Centers Cube is now set to On by default.
The default filter settings in the SpruceFilterCube Cube no longer flags for issues related to alternate locations as the toolkit warning is too strict and most structures can prep without issue.
The ReceptorInDUCube now has better handling and options for apo pockets.
The following improvements and modifications have been made to the Input PDB Codes for Spruce Cube.
PDB codes with only a CIF structure file may not find its associated MTZ file during prep.
The new Max File Size parameter will ensure downloads from the RCSB do not incur memory failures.
v0.27.0 February 2024
General Notice
This package is built using the following dependencies:
openeye-orionplatform>=5.1.1,<7.0.0.OpenEye-Toolkits>=2023.2.3.OpenEye-szmap>=1.6.7,<2.0.0.OpenEye-szybki>=2.7.0,<3.0.0.
The Cubes FastROCS Molecule Preparation and Parallel FastROCS Molecule Preparation have been removed from exports in the snowball and snowball.shape namespaces. They will need to be explicitly imported when used, that is, with
snowball.shape.frocs_prep import FastROCSDataPrepCube, ParallelFastROCSDataPrepCube.
New Cubes
The new Chomp Report Cube writes a Floe Report containing BROOD fragment database information.
Cube Updates
The ability to use random starts has been added to the ROCS, ROCS with Multiple Query Molecule, and ROCS for EON (Multiple Query) Cubes.
New parameters for warts to allow generation of unique conformer titles have been added to the following Cubes:
The Has Color Atoms Cube now ignores incoming records without a molecule field, instead of sending those through the failure port.
The Find ShapeQuery Cube now creates a new shape query corresponding to each conformer of a query molecule.
The Cubes BroodCube, BroodDBPacker, BroodDBUnpacker, BroodDBReader, BroodMatchCube, BroodHitListCube, and BroodDBMergeCube have been removed from Snowball.
The Cubes ChompFragmentationCube, ChompLibraryCube, and ChompMergeFragmentsCube have been removed from Snowball.
v0.26.1 September 2023
General Notice
This package is built using the following dependencies:
openeye-orionplatform>=5.1.1,<7.0.0.OpenEye-Toolkits>=2023.1.0.OpenEye-brood>=3.2.1.0,<4.0.0.OpenEye-szmap>=1.6.6.0,<2.0.0.OpenEye-szybki>=2.6.0.0,<3.0.0.
v0.26.0 July 2023
General Notice
This package is built using the following dependencies:
openeye-orionplatform>=5.1.0,<6.0.0OpenEye-Toolkits>=2023.1.0OpenEye-brood>=3.2.1.0,<4.0.0OpenEye-szmap>=1.6.6.0,<2.0.0OpenEye-szybki>=2.6.0.0,<3.0.0
New Cubes
The new BroodDBMergeCube Cube uses the
brooddbmergeapp to merge the 3D BROOD databases sent to its intake port to produce a merged 3D BROOD database.The new Find Target Name Cube ensures a target name is properly attached to an MMDS target record. If not found, the information will be inferred from header information.
A new cube, Brood DB Reader, has been added that reads contents of a BROOD database provided as a tarball and converts that into a set of records.
New cubes Brood DB Records Packer and Brood DB Records Unpacker have been added that packs sets of BROOD database records into shards and back.
A new cube, BroodMatchCube, has been added that performs a match between a BROOD database fragment against a given query.
A new cube, Brood Hit list, has been added that generates a hit list from a given set of BROOD matches.
Cube Updates
The following additional parameters are now available in Omega Conformation, Omega Conformation (Modes), Omega Conformation (ROCS mode), Omega Conformation (pose mode), and Omega Conformation (dense mode):
External fragments library
Fix based on maximum common substructure (MCS)
Nitrogen stereo enumeration
Ring enumeration
Flag to ignore stereo failures
Hydrogen sampling
The Enumerate Stereo Centers Cube can now handle enhanced stereo information.
The
Enumerate Nitrogenparameter in the Enumerate Stereo Centers Cube now defaults toFalse.The ChompMergeFragmentsCube Cube stores fragment information more efficiently to reduce its memory consumption.
The ChompLibraryCube Cube has been modified to generate conformers from the 2D fragments instead of working with the molecular conformers passed to the cube.
The Atom Potentials Cube fixed an issue with the atom potentials not being properly set on the output record.
The Torsion Scan Cube exposes the options for the “Penalty force constant” as well as the new “Number of Starts” and “Overlap divider between starts” torsion options which were added to Szybki TK in the 2023.1.0 toolkit release. This cube also has the option to calculate partial charges, in order to properly support the Parsley and Sage force fields.
The Bound Ligand Entropy Cube has been modified to take design units as input, and the cube can now work with
FF14SB-Sageand similar custom force-fields.The Ligand Entropy Cube has been modified to enable working with custom OpenFF force-fields.
The MMPBSA (Series) Cube now performs calculation on each of the conformers of a given ligand, when ligands with multiple conformers are passed.
The Freeform Ligand Strain Cube output records now contain both the
free ligandand therestrained ligandconformers corresponding to a tracked ligand conformer.
v0.25.3 December 2022
General Notice
This package is built using the following dependencies:
openeye-orionplatform>=4.5.4,<5.0.0OpenEye-Toolkits>=2022.2.2OpenEye-brood>=3.2.0.1,<4.0.0OpenEye-szmap>=1.6.5.1,<2.0.0OpenEye-szybki>=2.5.1.1,<3.0.0
New Cubes
Two new cubes Index Generator and Sorting Records by Index have been added that adds new field with integer indices and sorts based on indices respectively.
New cubes Interaction Hint Fingerprint Calculator Cube, and Receptor InteractionHint Fingerprint Annotation Cube have been added. These annotated docking results and receptor datasets, respectively, with fields that can be used for filtering in Orion based on interactions between a docked/posed molecule and the receptor. Additionally, an interaction fingerprint is calculated along with a Tanimoto score to the fingerprint of the ligand bound in the receptor as a reference (if available).
A new cube Spruce DU to Mol Simple has been added that subsets a design unit to desired components as a single molecule. A parallel version is also available.
A new cube Record to PDB File Converter has been added that converts a molecule to a PDB file and writes it to an Exports folder in the users “My Data” folder. A parallel version is also available.
Two new cubes Structure Standardization and DU Validator, and their parallel equivalents were added. The functionality for these cubes have been removed from Spruce Prep and divided into these separate cubes that are used before and after the prep cube. This is to more clearly denote where a structure fails in the standardization and preparation process. Parallel versions of these cubes are also available.
A Check for Partial Charges Cube was added that can check if an input molecule has been assigned partial charges. The record will be emitted through the success port if charges are present and through the failure port if not.
Cube Updates
New option
PBhas been added to thesolvent_modelparameter used in the following cubes.The max_value for the hit_list_size parameter in the _HitListBase cube has been increased to 100,000 records and the memory needed for the cube has been increased to support this larger number of records.
A bug was fixed in the Input Files for Spruce cube that caused a crash if part of the read structure did not have assigned chain IDs.
A build_loops boolean parameter was added to the Loop Database File Look-up cube.
The MMPBSA (Individual) amd MMPBSA (Series) cube now report the separate MM, PB, and SA scores along with the total MMPBSA score.
A bug in the Surface Area cube in ZAP and Polar Surface Area in SPICOLI, that left the surface area fields unpopulated/unchanged, was fixed
A bug in the _ConfValueHitList cube that didn’t access conformers correctly is fixed. This was causing the OEDocking to fail in certain cases when the
nposeswas > 1.The Atropisomerism was modified to include a check for the availability of partial charges for the molecule before checking for atropisomerism.
The cubes for energy calculation and optimization of a ligand in a rigid protein available in SZYBKI are modified to work with design units. The Protein Ligand Energy Individual and Fixed-Protein Ligand Minimization Individual can be used when a single ligand-protein pair in the DU is to be optimized. The Protein Ligand Energy Series and Fixed-Protein Ligand Minimization Series can be used when the multiple molecules (provided at the input port) are to be optimized within the same rigid protein (provided at the init port)
The Brood Query checks to see if the query molecule is 3D and fails otherwise
The Brood Fragment Selector allows for up to 50 atoms to be selected during fragment selection
The ChompFragmentationCube has lower and upper limits for all parameters where a range is applicable
The ChompMergeFragmentsCube has lower and upper limits for the minFrequency parameter
v0.24.1 September 2022
General Notice
This package is built using the following dependencies:
openeye-orionplatform>=4.4.0,<5.0.0OpenEye-Toolkits>=2022.1.1OpenEye-brood>=3.1.7,<4.0.0OpenEye-szmap>=1.6.4,<2.0.0OpenEye-szybki>=2.5.0,<3.0.0
Cube Updates
A bug with filter file input in the Molecule Filtering Using Custom Filter File cube was fixed.
The GetContextIDs MMDS cube has been improved to be able to handle much larger contexts.
The Unroll MMDS child-records into DU records fail-case has been improved to emit a log of the failure.
The Update Design Unit Style new check added to fatally end the floe if more than one query is submitted.
The Receptor in Design Unit cube added new parameter and check to better handle bioDUs.
v0.24.0 July 2022
General Notice
This package is built using the following dependencies:
openeye-orionplatform>=4.4.0,<5.0.0OpenEye-Toolkits>=2022.1.1OpenEye-brood>=3.1.7,<4.0.0OpenEye-szmap>=1.6.4,<2.0.0OpenEye-szybki>=2.5.0,<3.0.0
New Cubes
The new Macrocycle Filter Cube filters out records containing macrocycle molecules.
The new ChompFragmentationCube generates fragments from molecules using CHOMP.
The new ChompConfsChecker checks if user input 3D conformers are provided as a separate dataset.
The new ChompMergeFragmentsCube merges fragments and corresponding frequencies as generated from the ChompFragmentationCube.
The new ChompLibraryCube generates the BROOD Database.
The new cubes MMPBSA (Series) and MMPBSA (Individual) calculates single point MMPBSA score for ligands in the context of active site.
The new EON Score (Multi Query) and Zap Ligand Prep are variations of the EON Score and Zap Ligand Prep respectively, that expects both the
queryand thefitmolecules to be on the same record, and thus can handle working with multiple query molecules.The new EON Merge (Multi Query) merges eon records for multiple query eon calculations.
The new cubes ROCS with Multiple Query Molecule and ROCS for EON (Multiple Query) are variations of the ref:cube_RocsBestOverlayCube and ROCS for EON respectively that deals with multiple single conformer query molecules.
The new cube Add IDs and Generate Random Set and the corresponding parallel cube were added, which allows users to generate a random set of integers in parallel, for use in random splitting and selection.
The new cube, Parallel Random Select Number or Percent Cube, was added, which allows users, together with the ParallelRandomIntegerIdSetCube, to do efficient, parallel random splitting and selection.
The new cube Create A Custom Molecule Filter File was added, which allows users to create a custom filter file, which can be used with the OEMolProp OEFilter object, and can also run this filter in the same cube on optional molecule input.
The new cube Mol Title from String Field was added, which modifies the primary molecule field on a record by adding titles to that field using a user-selected string field on that record.
The new cube Multistate pKaModel-based Ionization States Enumeration enumerates the reasonable ionization state(s) of input molecules at neutral/physiological pH (7.4) based on pKa assessed using multistate pKa model.
A new cube Loop Database File Look-up has been added to lookup the loop database needed for loop modeling in protein preparation.
A new cube SiteHopper DB Prep has been added for reading a collection of design units an generating a SiteHopper Patch database.
The Bemis Murcko Scoring cube was added, which calculates a number for scoring Bemis-Murcko frameworks.
The Molecule Filtering Using Custom Filter File cube was added, which filters molecules using an OEMolProp custom filter file.
The Numeric Field To Constant Value Comparison cube was added, which filters records by comparing a numerical field to a constant value using a choice of near, not near, less than, less than or equal to, greater than, or greater than or equal to comparisons.
The Rename A Field cube was added, which renames a field passed to the cube.
The Decompose Similarity Matrix and Decompose Similarity Neighbor List cubes were added, which decompose NxN similarity matrices into smaller MxM sub-matrices using different methods.
The Fingerprint Generation (User-Defined) cube was added, which generates user-defined fingerprints for molecule records.
The Fingerprint Neighbor Similarity Calculation and Fingerprint Set Similarity Calculation cubes were added, which calculate fingerprint similarities using differently optimized methods.
Cube Updates
A new
solvent modelparameter has been added to the following cubes. The error handling behavior of these cubes have also been improved.ProteinLigandEnergyCube
FixedProteinLigandMinimizeCube
The Assign Ligand Partial Charges now properly handles 2D molecule inputs. The cube also now activates explicit hydrogen atoms irrespective of if the cube was assigning charges or if it was passing forward the available input charges.
The Posit now reports an additional field containing the
posit methodthat was used for a prediction. The cube also now properly passes records through thefailureport in cases when positfailsdue to low probability or clash as desired.The EON Score now keeps all the input record fields in the generated output record.
The Freeform Ligand Strain now keeps all the input record fields in the generated strains output record, along with having those fields being kept in the generated ensembles record.
The Overlap and Overlap with Grid Query cubes now prepares the input molecules with addition of color atoms and removal of hydrogen atoms, if the molecule did not have any color atoms.
The Find Protein cube was modified to add the found protein as a new field in the existing record instead of creating a new record.
The Torsion Scan has been improved to include a new force field parameter. The boolean parameter to allow using PB single point energies have been dropped in favor of a new solvent model parameter, that provides additional option to perform a gas-phase calculation.
The Spruce Prep has been improved to incorporate the new OESpruceFilter toolkit function, which standardize a structure read from a PDB or MMCIF file, but also fails the structure if specific problems are detected. An option is available to allow failures to pass. Additionally, the new OEValidateDesignUnit toolkit function is used to both style problematic areas of a design unit, and log messages to a validation floe report.
The Spruce Prep has been improved to no longer query for the loop database file, but now takes the file ID as initializer input from Loop Database File Look-up. This improves the cubes behavior at large scale use.
The Spruce DU to Mol has been improved, to properly mark the first generated molecule as the primary molecule on the record, making export of design units as molecules easier.
The Update Design Unit Style has been improved to fail early if the query record does not contain a valid design unit query.
The SiteHopper DB Prep and SiteHopper Search have been improved to store and retrieve, respectively, the PDB code, DU title, and target name if available with the data being used to build the database from.
v0.23.1 December 2021
General Notice
This package is built using
openeye-orionplatform>=4.2.4,<5.0.0andOpenEye-Toolkits==2021.2.0.
New Cubes
The new Field Type Conversion Cube converts a field of one basic type (integer, float, boolean, or string) to a field of another basic type.
The new String Field Comparison to Sets of Strings compares the value of a string field to a set of strings and routes records based on whether the value of that string field is contained in the set.
The new Process Every N records cube samples every N records from its input, emits those to its success, and others to the failure port.
The new Add hydrogens to design unit cube adds protons and optimizes the hydrogen bond network on the input design units or molecules.
The new Create DU From Pockets cube takes a design unit and searches for potential binding sites on the protein structure using OEPocket.
The new Create DU from f-pocket pockets cube takes a design unit and searches for potential binding sites on the protein structure using F-pocket.
The new ROCS Query Model cube builds OEShapeQuery model from bound ligands in the design unit(s)
The new Check Design Unit cube identifies if a record contains design unit field and emits the record through different ports accordingly.
The new Chemgauss4 Score (Individual) cube calculates checmgauss4 score between the ligand and receptor contained in the input design unit.
The new BINT Individual Score and Bint Series Score cubes calculate bint scores between the ligand and receptor contained in the input design unit.
Cube Updates
The following modifications have been made to the Posit cube:
Ability to generate multiple poses
Parameter to control multiple pose generation using only the best receptor or all the receptors.
Failed records to contain a pose when it fails due to clash or low probability.
The cube no longer calculate or add output record fields with chemgauss4 score or bint score
The Find ShapeQuery and ROCS were modified to allow working with custom color force fields.
New parameter to enable setting ionic state parameter was added to the following cubes:
The following ligand based cubes now provide a list of choices for built-in force fields, and also accept SMIRNOFF force-field XML files for custom force fields through a
BinaryInputPort. Additionally, new force fieldsage_openff200has also been added as a built in force field.The following protein-ligand based cubes now provide a list of choices for built-in force fields, and also accept SMIRNOFF force-field XML files for custom force fields through a
BinaryInputPort. For a custom SMIRNOFF input a newOEFF14SBSmirnoffinternally created. Additionally, new force fieldff14sb_sagehas also been added as a built in force field.ProteinLigandEnergyCube
FixedProteinLigandMinimizeCube
The output restriction energy records of the Freeform Conf Restriction Energies and Freeform Ligand Strain now contains the following additional fields:
Partner (ensemble) conformer index
Partner (ensemble) conformer relative energy
Partner (ensemble) conformer deltaG
The Spruce Minimize Design Unit Cube has been updated with fixes to the fixed and flexible selection, and now also allows full system minimzation.
The SiteHopper Search has been updated to use the SiteHopper Toolkits. The search cube also depends on the new SHDB toolkit file format stored in collections and older databases can no longer be used for search.
The SiteHopper DB Prep has been updated to generate a collection that can be used for search. Each shard in the collection contains a SHDB file that the SiteHopper toolkit uses to search.
Added option to process AlphaFold inputs for PDB codes to Records
Added option to include AlphaFold data to the Fetch all current PDB codes if tar file is specified
Added check to ensure AlphaFold structures will not be deleted in the Check for collection dupes
Reworked the GenerateGtoPFamilyTreeData tree to better reflect data and accommodate the inclusion of more PDB codes.
The GenerateGtoPTargetData has been updated to include Uncategorized (Non-GtoP) targets and keyed on their Uniprot ID protein name. Incorporation of AlphaFold into the tree based on Uniprot ID. Support of uniprot proteins that contain multiple targetable proteins. More stable internet queries. Human Ukbid ID are now searchable.
Improvements to the reference structure picker in DefineTargetReferenceStructure
Added SiteHopper superposition method to DefineFamilyReferenceStructure, and other code improvements to accommodate the additions of AlphaFold and Uncategorized proteins.
Spruce Prep for MMDS will now prepare a bio design unit of a structure without a ligand, and better handling of structures from uncategorized targets, as well as other minor bug fixes.
The Add Design Unit Field cube now has a parameter to make the additional first record out of this cube, containing the design unit, optional.
The Brood Report Cube now has an additional entry for query input, and uses the query molecule to set the size of the depicted molecules properly.
Deprecated Cubes
The following cubes have been deprecated and will be removed in a future release.
DatasetReaderOptCube. This cube is no longer relevant as the DatasetReaderCube in OpenEye-orionplatform has been updated to allow this functionality.
ShardReaderOptCube. This cube is no longer relevant as the ShardReaderCube in OpenEye-orionplatform has been updated to allow this functionality.
GPU SiteHopper Search. This cube should no longer be used as the toolkit does not allow search directly with a GPU, but only uses it as a pre-screening method.
v0.20.1 June 2021
Upgrades to
OpenEye-orionplatform==4.0.0and the associatedOpenEye Toolkits
New Cubes
The new Receptor in Design Unit Cube generates receptors inside design units.
The new ProteinLigandEnergyCube calculates energy for ligand conformers in a fixed protein active site.
The new FixedProteinLigandMinimizeCube minimizes ligand conformers in a fixed protein active site.
The new Flex-Protein Ligand Minimization minimizes ligand conformers in a flexible protein active site.
The new Add Design Unit Field adds a DU field to the first record of a set of records passed.
The Assign Ligand Partial Charges was created, which has the same functionality as the old LigandPrepCube.
The new Protein Preparation Report checks if the intake port record contains a log message field, and if so, retrieves and collects those and generates a report.
The new Protein Preparation Issues Detector checks the generated OEDesignUnit and adds log messages related to detected issues.
The new String Parameter to String Field Cube splits a string parameter into string fields on records, using an input separator.
The new Subset Router Cube routes the subset input to a different type of subset cube depending on the type of subsetting specified in the subset_type parameter.
The new Deduplicate Records Based on Integer, Float, String, or Mol Field deduplicates records based on a string field, a numerical field, and/or a mol field.
The new Deduplicate Records Based on Best Similarity Score deduplicates records based on a molecule field, retaining one record, for each molecule, with the best similarity score.
Cube Updates
The Find Receptor was updated to be able to find and emit a DU-receptor.
The FRED/HYBRID Exhaustive Docking Cube, and similar cubes, were updated to work with DU-receptors.
The Exception Handler was updated to avoid processing a fatal error log also being processed as regular failed record log.
The Posit was updated to check more thoroughly for input ligand molecules, and to work with DU-receptors.
The ROCS Receptor Selector Cube was updated to adjust to the switch to DU receptors.
The Omega Macrocycle was updated to not allow output of molecules with dimension less than three.
The Freeform Ligand Strain was updated to fix a bug that was producing negative local strain and global strain values.
The Posit was updated to output the Chemgauss4 Score for the posed molecule and the number of good interactions as measured by BINT.
The AddContexts cube was updated to skip adding targets to MMDS if another target with their same parent name alrady exists.
The Spruce DU to Mol cube was updated to use sensible defaults for field names. It was also updated to give users more control and the ability to output additional mol components into separate fields.
The Float Hit List cube, Integer Hit List cube, and String Hit List cube were updated to use their specific respective field parameter types.
A bug with the URL in the GenerateProasisFamilyTreeData cube was fixed.
The superposition method selection for families was fixed in DefineFamilyReferenceStructure cube.
The Input Files for Spruce cube was updated to properly read in .gz files.
v0.20.1 November 2020
Upgrades to
OpenEye-orionplatform==3.1.0and the associatedOpenEye Toolkits
New Cubes
New Atropisomerism Cube determines whether molecules have atropisomerism or not.
New Dipole Moment Calculation calculates dipole moment for input molecules.
New Molecule Counter Cube keeps counters for input molecules identified by the uuid field in the record.
New Success Counter Shard Cube, that counts shards. Behavior is identical to the parent Success Counter Cube.
A number of cubes have been added to support SiteHopper patch database generation and searching.
New SiteHopperPatchDBGenCube Cube that reads in a collection of prepared OEDesignUnits on records and creates SiteHopper patches from the binding site of each OEDesignUnit. A parallel version also exists ParallelSiteHopperPatchDBGenCube.
New SiteHopperPatchDBDatasetCube Cube that ties in the prepared OEDesignUnit collection with the collection of patches into a single record during patch databse generation. The SiteHopperUnpackDBDatasetCube Cube is used during search to extract the proper collection IDs for use by the search and related cubes.
New SiteHopper Search Cube, that performs a SiteHopper search on a supplied patch database using a reference OEDesignUnit as the query. A parallel version of this Cube also exists, Parallel SiteHopper Search. In addition a serial and parallel version that utilizes the GPU to search exists GPU SiteHopper Search and Parallel GPU SiteHopper Search.
New SiteHopperResultTransformation Cube, that performs a normal CPU re-scoring of the patches, as well as the option to normalize the patches against the query patch and re-score the similarly sized patches. The cube transforms the OEDesignUnits onto the reference frame, with the overlay provided by SiteHopper. A parallel version of this cube is also provided, ParallelSiteHopperResultTransformation.
New DUSequenceScore Cube, that compares the sequence of the SiteHopper query OEDesignUnit to the hits, with the ability to filter out results that are too similar in sequence. A parallel version of this cube is also provided, ParallelDUSequenceScore.
New Update Design Unit Style Cube, that colors the OEDesignUnits for each hit in the output records with random colors to ensure easier visualization in 3D.
A number of cubes have been added to support populating MMDS using Floe. This includes generating a collection of prepared OEDesignUnits, datasets of targets and target families as well as cubes that allow uploading data into MMDS (for MMDS admins only).
New Create Structure Collection Cube that either creates or updates an existing collection. The Cube emits the collection and existing shards via different ports.
New Fetch all current PDB codes Cube that retrieves all known PDB codes from the RCSB and makes a record for each with the code and the latest revision.
New Check for collection dupes Cube that reads all the shards in a provided collection and checks whether a PDB code and revision already exists in the collection. Only codes not already present are emitted. Shards that are present but where the code has been deprecated or a new revision has been added are deleted from the collection.
New PDB codes to Records Cube that works similarly to the existing Input PDB Codes for Spruce Cube, but instead of input parameters takes input from records provided via the intake port. A parallel version of this cube is also provided, Parallel PDB codes to Records.
New Convert Structure Records to shards Cube that takes records with PDB data and converts them into a shard each. The shard is named by the PDB code for easier look-up later and the PDB revision (ordinal) is stored in the shard metadata. A parallel version of this cube is also provided, Parallel Convert Structure Records to Shards.
New GenerateGtoPFamilyTreeData Cube that reads the family hiearchy from the Guide to Pharmacology database and generates a dataset from it.
New GenerateGtoPTargetData Cube that reads the target information from Guide to Pharmacology and maps the target UniProtKb IDs to PDB codes.
New UpdateTargetStructures Cube that can update a dataset of targets if new structures have been added to that target since last updated.
New DefineTargetReferenceStructure Cube that attempts to find a common reference structure for a given target based on a consensus biological unit and binding site location. A parallel version of this cube is also provided, Parallel Define Target Reference Structure.
New DefineFamilyReferenceStructure Cube that reads the reference structures for each target in a family and looks to see if they can all be superposed to a common reference frame. It subsequently checks whether each family member node can be superposed with other family members at the same level in the tree.
New TargetStructureMappingCube Cube that takes a dataset of targets and matches up the reference structure for a given target to each of the PDB codes belonging to that target for structure preparation. A parallel version of this cube is also provided, Parallel Target Structure Mapping.
New Spruce Prep for MMDS Cube that works identically to the existing Spruce Prep, but instead of records operates on shards. An output shard contains all the prepared OEDesignUnit records for a given PDB code. A parallel version of this cube is also provided, Parallel Spruce Prep for MMDS.
New Spruce depiction cube for MMDS Cube that works identically to the existing Spruce Depiction, but instead of records operates on shards. A parallel version of this cube is also provided, Parallel Spruce Depiction Cube for MMDS.
New AddFamilies Cube that takes a dataset and builds a family tree from it.
New AddContexts Cube that takes a dataset and adds every context (target) at the proper location in the family tree. A parallel version of this cube is also provided, Parallel Inject Target Tree into MMDS.
New AddExperimentsAndSites Cube that reads a collection of data for each PDB code and merges it into MMDS. It also reads a collection of prepared OEDesignUnits and uploads these as well. A parallel version of this cube is also provided, Parallel Inject Experiment and Site into MMDS.
New AddReceptorForFrame Cube that adds OEDocking receptors to every frame in MMDS. A parallel version of this cube is also provided, Parallel Add receptor for a frame in MMDS.
New AddContextSeqAlign Cube that pulls all the frames for each context and generates a sequence alignment for all the structures. Contexts with an existing alignment are skipped, unless the context has had new structures added. A parallel version of this cube is also provided, Parallel Add Sequence Alignment to a Context in MMDS.
New GetContextIDs Cube that outputs a dataset of existing context (target) IDs from MMDS.
New GetFrames Cube that outputs a dataset of existing frame IDs from MMDS.
A number of cubes have also been added that instead of reaching out to public sources instead can extract data from a Proasis server based on a user provided host url.
New GetProasisCodes Cube, mimics Fetch all current PDB codes. A specialization of this cube has also been added that instead of reading all existing codes just reads the latest codes based on a number of days in the past, GetRecentProasisUpdates Cube.
New ProasisCodesToInputRecords2 Cube, mimics PDB codes to Records Cube. A parallel version of this cube is also provided, Parallel Structure IDs to Records.
New GenerateProasisFamilyTreeData Cube, mimics GenerateGtoPFamilyTreeData.
New GenerateProasisTargetData Cube, mimics GenerateGtoPTargetData.
New DeletePDBCodesCube Cube that removes a bad PDB code from MMDS, the prepared OEDesignUnit collection and the PDB file collection.
Cube Updates
The Input PDB Codes for Spruce now adds the revision number (ordinal) when pulling data from the RCSB.
The Assign Partial Charges Cube was improved. A parameter was added to specify whether to use input conformers or not for am1bccelf10 charges. If not, Omega will be used to generate conformers.
The Gameplan was updated to handle failures to the fatal port.
The Exhaustive Docking (Single Receptor) no longer emits a record with emtpy molecule when the docking produces no results.
The Freeform Solvation Energy was updated so that the output molecule from the solvation is on the output record. Parameters were also added for chosing the ionic states for the calculation and if the input 3D structure should be solvated without further optimization.
v0.19.3 August 2020
Cube Updates
The Spruce Depiction was updated to handle modeled structures where Iridium data is not available on the DesignUnit
The Spruce Prep was updated to resolve issues with modeled loops.
v0.19.1 August 2020
Upgrades to
OpenEye-orionplatform==3.0.0the associatedOpenEye Toolkits
New Cubes
New Spruce Depiction which generates multiple interactive depictions of the design unit. The depictions are target-ligand interactions, b-factor visualization, 3) Iridium data visualization, and if electron density data is available, 4) ligand density coverage.
New DesignUnit Type ETL. OEDesignUnit have gotten native support in datarecord as Types.Chem.DesignUnit. This cube converts fields with the old design unit blob type and adds a new field with the native type. For backwards compatibility it currently preserves the existing field.
New Named Dataset Reader, Named File Reader, and Named Collection Reader read dataset, file and collection specified by their names. They allow floe developers to permenently customize their floes to utilize a specific resource.
New Calculate RMSD performs RMSD calculation between a 3D reference molecule and multi-conformation molecules.
New Find ShapeQuery identifies shape query in a record. If a shape query is not found, the cube looks for the primary molecule and creates a shape query from that molecule.
New Convert Shape Query to Record reads a shape query file (.sq) and converts the shape query to a record.
New Single File Reader reads a single file and emits its contents in a stream. It will throw a runtime error if two or more files are used as input.
New Record Splitting By Charges splits records into two streams by whether the molecule is charged.
Cube Updates
The Tautomer Enumeration was updated to expose more tautomer options. The enumerated tautomers are now also set to the primary molecule on the output record.
The Merging Conformers was updated to handle missing children failures without raising exceptions.
The Spruce Prep has been updated to incorporate changes in the recently release Spruce Toolkit, as well as added support for loop modeling, which is on by default. Loop modeling can add considerable time.
The Find Protein has been modified so the user optionally can specify, the molecule field to use. If the protein is extracted from a design unit, the user now has the option to get other components of the system along with the protein, e.g. cofactors and/or solvent. The cube now also does a minimal check that the molecule extracted is a protein, based on number of standard protein residues. The logic has changed such that the molecule of interest, is emitted on a new record with the molecule as the primary molecule.
The Find Ligand has been modified so the user optionally can specify, the molecule field to use. The logic has changed such that the molecule of interest, is emitted on a new record with the molecule as the primary molecule.
The Activesite Ligand Minimization and Ligand Minimization no longer throw fatal errors when PB solvent mode is set to PB.
The Convert Conformers to Mols output records that no longer contain all conformer records.
The Assign am1bcc partial charges now generates correct charges.
The Brood Report Cube has been improved to align depictions of hits with the non-changing part of the BROOD query.
The Omega Conformation now supports fix molecule and fix smarts to fix an portion of the input molecule.
Support of Open FF 1.0 has been added to the following cubes. PARSLEY_OPENFF100, PARSLEY_OPENFF111 and PARSLEY_OPENFF120 are added as predefined choices, and an .offxml file that defines a SMIRNOFF force field can also be used as input.
The ROCS, SubROCS, and Best Overlay Transform now take a record of shape query from init port. In order to use a molecule as query, Find ShapeQuery can be used to create a shape query from the molecule.
The Record Splitting By Conformers and Record Splitting By Initializer Conformers have been updated to handle shape query in addition to molecule.
Removed Cubes
Removed DUFileToRecords cube which is no longer needed as Orion will automatically, upload and ETL an OEDU file to a record.
v0.18.2 April 2020
General Notice
Upgrades to
OpenEye-orionplatform==2.4.5
New Cubes
New Record Counter Switch added that can switch floe path based on the number of records in the initializer port.
New Rotate Surface rotates a surface by a 3x3 matrix specified as a 9-element array in row-major order.
New Transform Surface transforms a surface using a supplied OETrans object.
New Translate Surface translates the surface using the supplied 3-element translation vector.
New Dataset Reader Optional to allow a user to pass an optional reference design unit to use when preparing new structures in Spruce.
New Input Files for Spruce, which is a re-work of the older and removed
PDBFileToInputRecord. This cube takes a set of input PDB files, optional MTZ files, and optional metadata JSON files and pairs them up as input records for Spruce structure preparation.New Input PDB Codes for Spruce, which is a re-work of the older and removed
PDBCodeToInputRecords. This cube downloads PDB (fall-back to MMCIF) files and any associated MTZ files and puts them into records for Spruce structure preparation.New Molecule Filtering cube has been added that filters molecules based on a user-defined filter type.
New Uncolor Molecule Graph cube has been added that uncolors molecular graphs i.e. removes certain atom and bond properties.
New Random N Record Selection cube has been added that randomly selects N records.
New Concatenate Records cube has been added that concatenates datasets.
New Classification Based on String Field cube has been added that allows classification of records based an a specified string field.
New String Fields Comparison to Sets of Strings cube has been added that filters out records based on the string data of reference records.
The following cubes have been added to mimic some of the stages of biomolecule preparation being done automatically in Spruce. They all, except the Spruce BioUnit Extract, take a dataset with designunits as input. The editing cubes updates the design units on the record. To be able to use them for downstream cubes or floes, after editing, the design units on the records must be converted to molecules or receptors (see conversion cubes section following this section).
New Spruce BioUnit Extract, which generates biological units from an input asymmetric unit crystal structure.
New Spruce sidechain builder cube, takes a dataset of designunits and builds out partial or missing sidechains.
New Spruce capping cube, takes a dataset of designunits and caps any chain breaks.
New Spruce swamp metal cube, takes a dataset of designunits and converts specified metals, e.g. Zn to Mg.
New Spruce mutation cube, takes a dataset of designunits and attempts to mutate specified residues.
New Spruce residue state cube, takes a dataset of designunits and changes the state of specified titratable residues.
New Spruce rotamers cube, takes a dataset of designunits and returns multiple records per design unit each with different rotamers of a specified residue (non-clashing is default, but optional).
New Spruce subset cube, takes a dataset of designunits and subsets them based on inputs, to remove entire design unit components or remove pieces specified by residue identifiers.
New Spruce subset within cube, takes a dataset of designunits and subsets a component of each design unit, e.g. subset the solvent to include only those within 5A of the protein and ligand components.
New Spruce Superpose, takes a dataset with a reference design unit and a dataset of designunits, and superposes each design unit in the dataset to the reference. Several superposition methods are available.
New Spruce Minimize Design Unit Cube, takes a dataset of designunits and minimizes parts of the designunits based on input parameters.
The following cubes have been added that convert design units to molecules or receptors, for use in subsequent cubes or floes.
New Spruce DU to Mol, which converts the design unit to a molecule and optionally splits out the ligand to a separate field. All the content in the design unit is output to the molecule, if less is required, first subset the design unit with the cubes: Spruce subset cube or Spruce subset within cube.
New DUToReceptor, is a cube split out from the old DUtoReceptorMDRecord, to split the two operations therein. This cube takes a DU and writes a receptor for use in docking.
New DUToMDRecord, is a cube split out from the old DUtoReceptorMDRecord, to split the two operations therein. This cube takes a DU and writes an complex (not including the ligand), that can be used in MD floes, e.g. Short Trajectory MD floes.
Cube Updates
The DUFileToRecords, has been updated to take multiple input files.
The Spruce Prep, has been updated, to take an optional reference design unit using the initializer port, additional options for preparation have been exposed, and several of the input parameters have been updated to make parameter selection easier.
The MDL Substructure Map now emits unmatched molecules through the failure port.
The Exhaustive Docking (Single Receptor) now exits properly when initialization fails.
The Torsion Scan now has a torsion increment option to change the resolution of the torsion drive. It also fails more gracefully when coordinates cannot be generated or the torsion cannot be identified.
The Splitting Conformers now has the option to move conformer fields to the primary record after splitting.
The Prepare Molecule for 2D Depiction cube now can keep the coordinates of the primary molecule and store the 2D coordinates in a new field specified by a new string parameter.
The following changes have been made for the Move Field cube:
It now handles cases when the specified field does not exist on the record.
A new parameter has been added that allows to move records with missing field to the success port.
A fatal port has been added in order to handle cases when no field was renamed in any of the records.
The following changes have been made for the Structure to SMILES cube:
Meta.Hints.Chem.SMILES meta data is added to the SMILES field
It re-perceives all necessary perception upon request.
It noew generates the following SMILES: non-canonical, non-isomeric-canonical, and isomeric-canonical (default)
Removed Cubes
Removed DUtoTgtLigRecords cube which was unused.
The following cubes have been removed and replaced with the new Input PDB Codes for Spruce:
GetPDBplusMap,PDBCodeToUrl,UrlToFile,PDBCodeToInputRecord,MultiplePDBCodesToInputRecord.The following cubes have been removed and replaced with the new Input PDB Codes for Spruce:
PDBFileToInputRecord,InputRecordtoDataset.The following cubes have been removed and replaced with new DUToReceptor and new DUToMDRecord:
DUtoReceptorMDRecord,ReceptorMDRecordToDataset,DUtoReceptorMDDataset.
v0.17.2 November 2019
General Notice
Upgrades to
OpenEye-orionplatform==2.2.0Upgrades to 2019.Oct release of the OpenEye Toolkits
New Cubes
New Ligand Energy to combine functionality of the
GasPhaseLigandEnergyCube,SheffieldLigandEnergyCubeand thePBLigandEnergyCube.New Ligand Minimization to combine functionality of the
GasPhaseLigandMinimizeCube,SheffieldLigandMinimizeCubeand thePBLigandMinimizeCube.New Ligand Restrained Minimization to perform free ligand minimization with an additional harmonic potential restraint on ligand atoms
New Activesite Ligand Restrained Minimization to perform activesite ligand minimization with an additional harmonic potential restraint on ligand atoms
New Ligand Hydrogen Optimization to adjust ligand hydrogen coordinates, while keeping the position of the heavy atoms fixed.
New Find Protein to be able to obtain protein from either a molecule dataset or a design units dataset.
New Find Ligand to be able to obtain ligand molecule from either a molecule dataset or a design units dataset.
New Gameplan Depiction to be able to generate 2D depictions of Gameplan results.
New Gameplan Report Cube.
New Record Splitting By Conformers to be able to sort out single and multi conformer molecule records.
New Record Splitting By Initializer Conformers to be able to redirect records through separate paths based on an initializer molecule record containing a single and multi conformer molecule record.
New 2D Fragment Picker to be able to select a fragment from a 2D sketched molecule.
New 2D Torsion Picker to be able to select a torsion fragment from a 2D sketched molecule.
New MDL Query Editor to be able to sketch an MDL query.
New Brood Fragment Selector to be able to select a Brood fragment in a molecule.
New MDL Substructure Map to be able to find substructures against MDL query.
New Brood Query to build Brood Query from molecule with selected fragment.
New Brood Report Cube to generate a report of successful Brood calculation.
Cube Updates
Activesite Ligand Energy have been enriched with the following functionality:
Ability to optimize in PB solvent. Accordingly,
PBProteinLigandEnergyCubehas been removed.Options for multiple force field choices
Improved validation and error handling, including the addition of a
fatalport for fatal errors.
Activesite Ligand Minimization have been enriched with the following functionality:
Ability to optimize in PB solvent. Accordingly,
PBProteinLigandMinimizeCubehas been removed.Options for multiple force field choices
Improved validation and error handling, including the addition of a
fatalport for fatal errors.
Ligand Torsions Minimization has been enriched with following functionality:
Options for multiple force field choices
Additional flag to turn on/off optimization, to allow single point energy calculation.
Improved validation and error handling, including the addition of a
fatalport for fatal errors.
EON Score has been enriched with following functionality:
Ability to calculate EON tanimoto combo.
Sort the input conformers based on tanimoto combo, and return only the highest scoring conformer.
Improved validation and error handling, including the addition of a
fatalport for fatal errors.
LigandPrepCube has been enriched with ability to choose a partial charge model, as well as with improved validation and error handling.
Zap Ligand Prep has been modified to always assign
MMFF94charges, as well as with improved validation and error handling.ROCS for EON has been modified to restrict its scope to a single conformer query molecule as well as with improved validation and error handling, including the addition of a
fatalport for fatal errors.Posit has been modified to use the capabilities of the newly modified Posit toolkit.
Omega Macrocycle has been re-implemented with the toolkit functionality, and accordingly snowball no longer depends on the
Omega App.To improve usability, cubes Freeform Conf Restriction Energies and Freeform Ligand Strain have been modified to emit restriction energies as separate records through a different port compared to the free energies records.
The following cubes have been modified to improve error handling:
The following cubes have been modified to correct functionality issues:
The following
OMEGAcubes have been modified to turn on canonical ordering of atoms by default:The following cubes have been removed due to their suboptimal usability
GasPhaseLigandEnergyCubeSheffieldLigandEnergyCubePBLigandEnergyCubeGasPhaseLigandMinimizeCubeSheffieldLigandMinimizeCubePBLigandMinimizeCubePBProteinLigandEnergyCubePBProteinLigandMinimizeCubeActiveSiteRstrLigandMinimizeCubeActiveSiteRigidLigandMinimizeCubeActiveSiteFlexLigandMinimizeCubeFlexProteinDistLigandMinimizeCubeSingleReceptorPositCubeFreeformEneStrainCube
v0.16.6 September 2019
General Notice
Upgrades to
OpenEye-orionplatform==1.1.4Upgrades to
OpenEye-floereport==0.1.5Static variables are no longer used, other than as the
parametervariables.
New Cubes
New Flipper to either enumerate stereo or not.
New Omega Conformation (Modes) to run omega in predefined mode.
New Failure Report that can produce a report of log information from failed records.
New Exception Handler to handle exceptions from downstream cubes and stop a flow.
New Success Counter to count pass-through records and report floe failure when connected properly to an Exception Handler.
New Find Receptor to find receptors in a record.
New Add Query Style to add viewing style to a query molecule.
Cube Updates
The following cubes have been enriched by addition of a
fatalport. Any failure during initialization is reported by emitting a record containing failure log information through thefatalport.SingleReceptorPositCube
OEStructureBasedLigandDesignCube
The following cubes has an additional flag that allows to control the amount of output results to be produced.
Removed
modeoption from Omega Conformation, to simplify it as a customizable cube.OEStructureBasedLigandDesignCube and SingleReceptorPositCube now properly reports the output
receptorand thebound ligand.Conformer Integer Hit List, Conformer Float Hit List, and Conformer String Hit List now works on the molecule record field if molecule has a single conformer and the conformer record does not contain the suggested field value.
DUtoReceptorMDDataset now has a new MD component mask and produces robust datasets for MD applications. Also, a small modification was made in how receptors are stored on the output dataset so that grids and other receptor-specific information can be displayed in the 3D window.
EStructureBasedLigandDesignCube and SingleReceptorPositCube now generate a single pose when relaxation is turned off.
v0.16.0 July 2019
General Notice
Upgrades to
OpenEye-orionplatform==1.0.0
v0.15.0 June 2019
General Notice
Upgrades to
OpenEye-orionplatform==0.3.2
v0.14.0 June 2019
General Notice
Upgrades to
OpenEye-orionplatform==0.2.5and the 2019.Apr release of the OpenEye Toolkits
New Cubes
New Gameplan Report Cube to report Gameplan results and app generated log files.
New Brood Report Cube to report Brood results and app generated log files.
New ROCS with Multi-conformer Query cube that works with a multi conformer query molecule.
New SingleReceptorPositCube specifically for single receptor pose prediction.
New DUtoTgtLigRecords to extract target and ligand DataRecords from OEDesignUnits.
New DatasetReaderOpt to read records with an optional data source.
Cube Updates
Exhaustive Docking (Single Receptor) no longer produce docked pose with unusually high docking scores.
ROCS, SubROCS and Best Overlay Transform now works with a single conformer query molecule only and no longer produce a Query Conformer field in the output record.
OEStructureBasedLigandDesignCube now checks for memory limits corresponding to the number of receptors and poser cash size before starting to pose.
Updated BroodCube documentation.
Updated Gameplan documentation.
Spruce cubes have been updated to use and output OEChem::OEDesignUnit object files (.oedu), which are new to the 2019.Apr release of the OpenEye Toolkits.
v0.13.6 March 2019
General Notice
Upgrades to
OpenEye-orionplatform==0.1.14
New Cubes
New Splitting Conformers and Merging Conformers cubes which preserve fields added during calculations on the split conformers.
previous cubes for Splitting and Merging conformers which did not preserve fields have been removed.
New Molecule Title To Field cube to create a field with the title of the molecule in a chosen MolField.
New Gameplan added that performs Gameplan calculations.
New Build conformer using Distance Geometry added that generates molecule conformers using distance geometry.
New Macrocycle Convergence added that along with the Build conformer using Distance Geometry can be used to create an efficient Macrocycle Omega floe that can generate macrocycle conformers in parallel mode.
New Omega Conformation (FastROCS mode) added that generates conformers for FastROCS.
New Align Conformers by RMSD Overlay added that can be used to align all conformers of a molecule based on RMSD.
New Omega Conformation (FastROCS mode) added that generates conformers for FastROCS.
New Copy Record added that can be used to generate multiple copies of a record.
New Torsion Scan that identifies torsions on each input molecule and generates copies of the record with a different torsion labeled by an atom tag on each copy.
New Activesite Ligand Energy added that can be used to calculate intermolecular energies between a ligand and a protein.
New BroodCube added that performs Brood calculations.
Cube Updates
Added new parameters mode and rotor_offset in the Omega Conformation.
Fixed a parameter mismatch issue with the Assign Partial Charges.
The Torsion Scan is updated to use a single tag to identify a torsion, instead of four tags. It also now has options to generate conformations using Omega prior to torsion driving or use a provided conformer ensemble. By default the cube reports both the MMFF/Sheffield energy as well as the MMFF/PB energy for each output conformer.
Fixed Zap Ligand Prep to fail gracefully when it cannot prep a molecule.
The Torsion Scan is updated to use a signle tag to identify a torsion, instead of four tags.
New parameters in_strain_mol_field and out_strain_mol_field added to the FreeformStrainCube and the FreeformEneStrainCube cubes to enable using separate input conformers for freeform free energy and strain calculations.
New parameter use_inp_ens have been added, to enable using user provided conformer ensemble in calculations, to the following freeform cubes:
FreeformConfEntropyCube
FreeformEneStrainCube
v0.13.4 November 2018
General Notice
Upgrades to use
OpenEye-orionplatform==0.1.9and the 2018.Oct release of the OpenEye Toolkits
Cube Updates
Minor fix to Omega Conformation to preserve molecule titles.
Speed up fix for many of the FreeFormConf cubes.
Updated the Hermite shape cubes for compatibility with the 2018.Oct release of the OpenEye Toolkits
v0.13.3 September 2018
General Notice
Upgrades to use
OpenEye-orionplatform==0.1.7
New Cubes
The following serial cubes have been added to merge fields from two records:
The following serial logic cubes and their corresponding parallel version have been added to compare on string fields:
Cube Updates
CalculateXLogPCube now calculates XLogP on the neutral form of the molecule. A new parameter has been added to the cube, called Keep Neutral Form, which controls whether to keep the neutral form of the molecule after the calculation or restore the original input form.
v0.13.0 August 2018
General Notice
Upgrades to use
OpenEye-orionplatform==0.1.0
Legacy Release Notes
v2.1.2 September 2025
This package is built using
OpenEye-orionplatform==6.5.1,OpenEye-toolkits==2025.1.1, andOpenEye-Snowball==0.30.2.
v2.1.1 August 2025
General Notice
This package is built using
OpenEye-orionplatform==6.5.1,OpenEye-toolkits==2025.1.1, andOpenEye-Snowball==0.30.0.
Floe Updates
The following improvement has been made to the OMEGA - 3D Conformer Ensemble Generation Floe: A new Conformer Generation Mode option, ROCS-TS, has been added which uses Thompson Sampling to generate conformers suitable for ROCS calculations.
v2.0.1 February 2025
General Notice
This is a new package as a part of the Orion Floes reorganization. It includes floes previously found in the Classic, Cheminfo, and Biomodeler Floes packages.
This package is built using
OpenEye-orionplatform==6.3.1,OpenEye-toolkits==2024.2.1, andOpenEye-Snowball==0.29.1.
Floe Updates
The following improvements and modifications have been made to the SPRUCE - Protein Preparation Floe:
Metadata handling has been improved. Metadata file input is promoted with improved descriptions and better logging if a metadata file was not matched to an input structure file.
The Add Ligand Smiles parameter has been added to better identify global ligand metadata and tautomer states.
The handling of receptors for covalently bonded ligands has been improved.
MTZ data handling has been updated after recent changes to the RCSB offerings of the EDMAPS Announcement.
The Calculate Average Precision Floe has been removed.