Tutorial: Library Enumeration for Hit Identification
This tutorial covers the following:
Creation of a reaction and reagent database
Inspection of the database
Product enumeration
We will use the following floes:
Tutorial Overview
In this tutorial, we will demonstrate how to enumerate the products of a given reaction using compounds sourced from a vendor database as potential reagents. The resulting enumerated library will be generated as a collection, with the option to export it as a SMILES file. For hit identification purposes, the enumerated library can be used with the ChemInfo Hit ID Floes package to create a 2D library suitable for similarity or substructure searches. Alternatively, it can be processed using the Large Scale Floes to generate a 3D collection for use with FastROCS or Gigadock Floes.
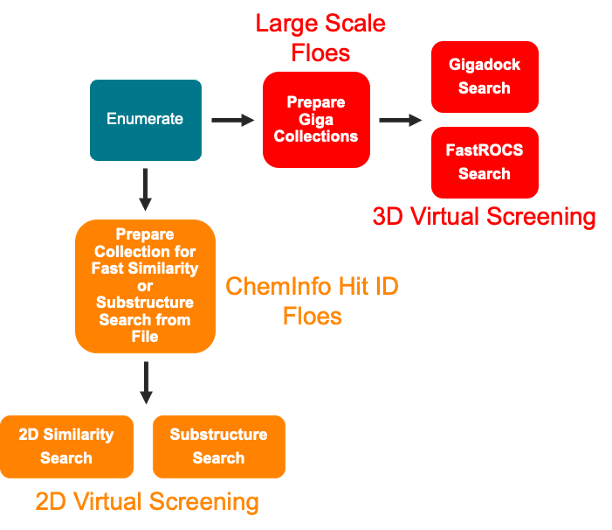
Figure 1. Workflow pipeline from enumeration to virtual screening.
How to Create and Inspect the Reaction and Reagent Database
The reaction and reagent database is an essential part of the workflow. It contains a set of building block structures that have been indexed for particular transformations, using logic found in the reactions and reagents definitions file. For hit identification and lead optimization applications, users are strongly encouraged to index their own reaction and reagent database, either using their own in-house collection building blocks or using vendor catalogs of specific interest to them. This will lead to the generation of ideas that are actionable as output down at the end of the process.
The default databases that are included in Organization Data on Orion are “sample” databases that allow users to explore the generative design floes while also being small and curated enough to include some reactants that can be used in supported transformations. If using these databases, you should expect only a few ideas, not a dataset of compounds that exhaustively generates possible analogs.
For this tutorial, we will enumerate the products of a Schotten-Baumann reaction using a library of amines and carboxylic acids given in separate datasets, each containing 5,000 molecules. The amines and carboxylic acids were sourced from the Mcule in Stock subset of the Mcule database (downloaded in May 2025 from the vendor website https://mcule.com/database/ ). These molecule files are smaller subsets prepared specifically for this tutorial and can be downloaded below.
The first step of the enumeration is to index the building blocks based on the logic of a reaction definition file. The reaction definition file provided in the Organization Data contains the rules to classify whether the reagents are compatible with a chosen transformation. For instance, if an amine contains a functional group incompatible with the Schotten-Baumann reaction, such as an aldehyde, it will be excluded from the enumeration. If you would like to make changes to the reaction definition file to allow incompatible functional groups, change a reaction definition, or even add a new reaction, please contact support at support@eyesopen.com.
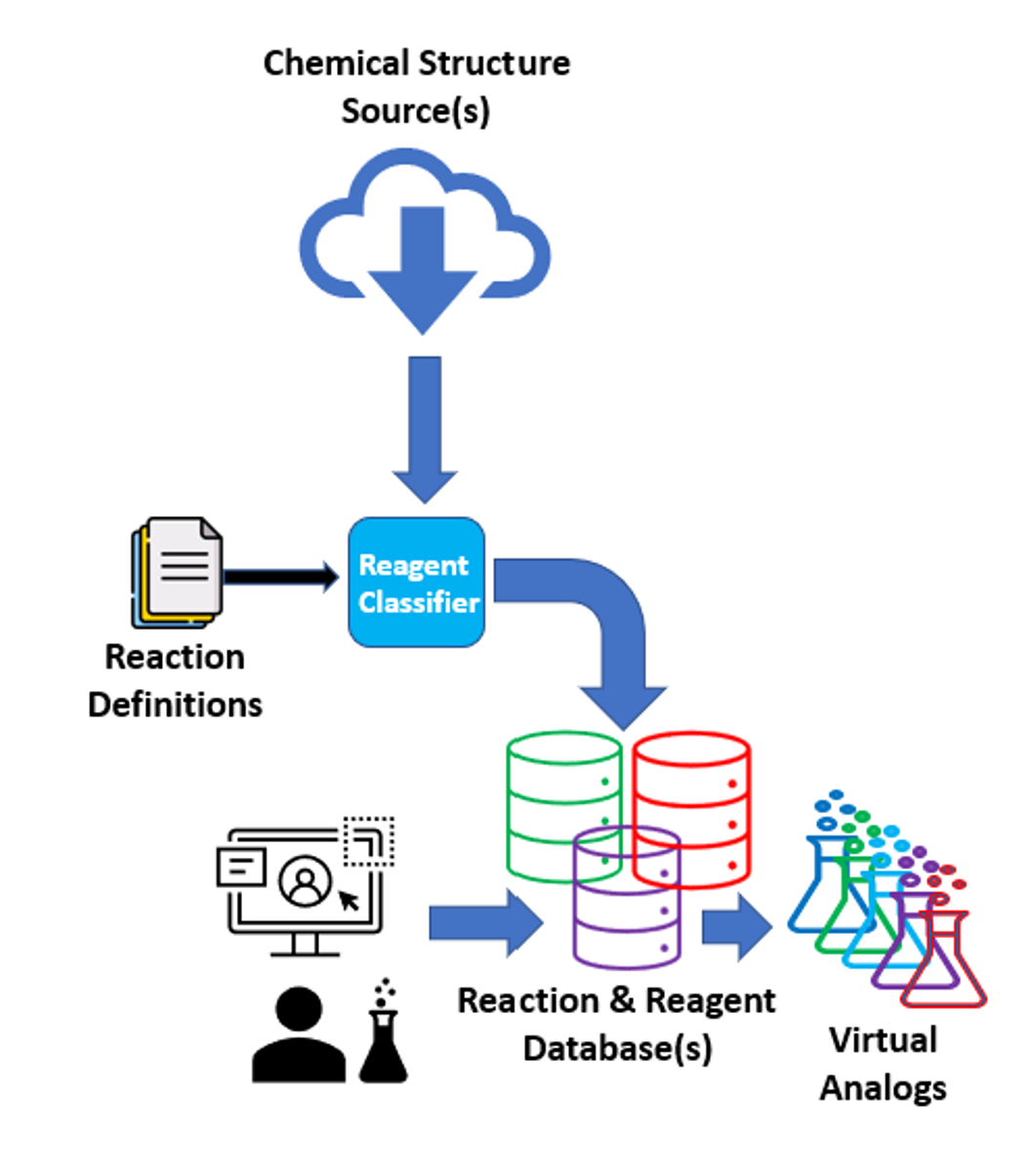
Figure 2. Schematic of the creation of a database with classified reagents and downstream processing (enumeration, virtual analogs, etc.).
Create the Reaction and Reagent Database
First, click the green “Add Data” button on the Data page. Upload the amines.smi and carboxylic_acid.smi files to Orion. An ETL floe will automatically convert the files to datasets. The dataset format allows you to inspect the library in the 3D & Analyze page. If you are reproducing this tutorial with a database containing more than 100,000 molecules, you must disable the automatic file-to-dataset conversion in the “Upload to Orion” window. Click on “Show Advanced Options” and choose None for the Processing Method (Figure 3).
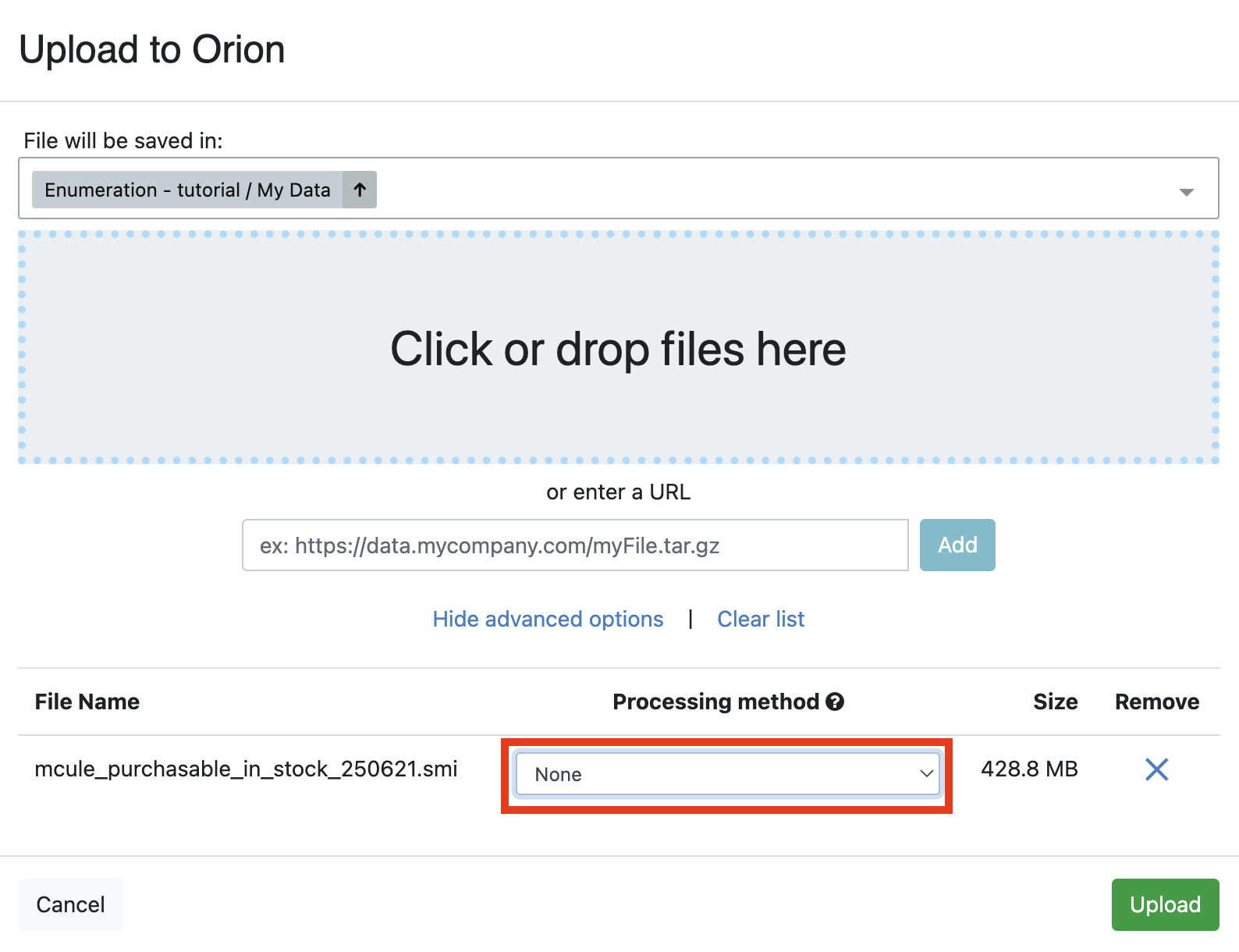
Figure 3. The “Upload to Orion” window with the option None selected for the Processing Method. The file uploaded in the example is the entire Mcule in Stock library.
On the Floes Page, type Reaction & Reagent Database into the search bar to locate the Reaction & Reagent Database – Create from Dataset Floe. Please note that if you are reproducing this tutorial with a SMILES file, you should use the Reaction & Reagent Database - Create from BULK SMILES Floe instead. For more information, please see the Create a Reaction and Reagent Database section of the tutorial for that floe.
Choose the Reaction & Reagent Database – Create from Dataset Floe. Click “Launch Floe” to bring up the Job Form. The parameters can be specified as below.
Input Parameters
Reaction Definition File: This file resource can be found in Organization Data on the Orion Data page, using this path: Organization Data / OpenEye Data / Generative Design Data. Select the most recent reaction classification file. At the time of this writing, the applicable file was
2025_1_sample_reaction_classification.txt.Input Dataset: Choose either the amine or carboxylic acids dataset. Click the “Add More” button to input the other reagent dataset.
Output Reaction & Reagent Database Name: Give the output database a recognizable name.
Dataset Processing Options
Molecule Field: Choose Molecule from the drop-down selections.
Reagent ID: Use the keyword Title.
Functional Group Transformation: Turn this Off. If On, the floe may generate new building blocks on the basis of simple functional group transformations, creating inaccessible analogs. For example, an ester can be converted to a carboxylic acid, resulting in a new molecule not listed in the catalog.
Salt Chop: Turn this On to ensure that you will use the largest (and most relevant) fragment in a salt.
Neutralize Charges: Turn this On. If there are charges in some building blocks (e.g., benzoxonium chloride), it is advisable to neutralize such charges to facilitate the functional group recognition.
Check that the parameters in your Job Form are as shown in Figure 4, then click “Start Job” to begin the floe.
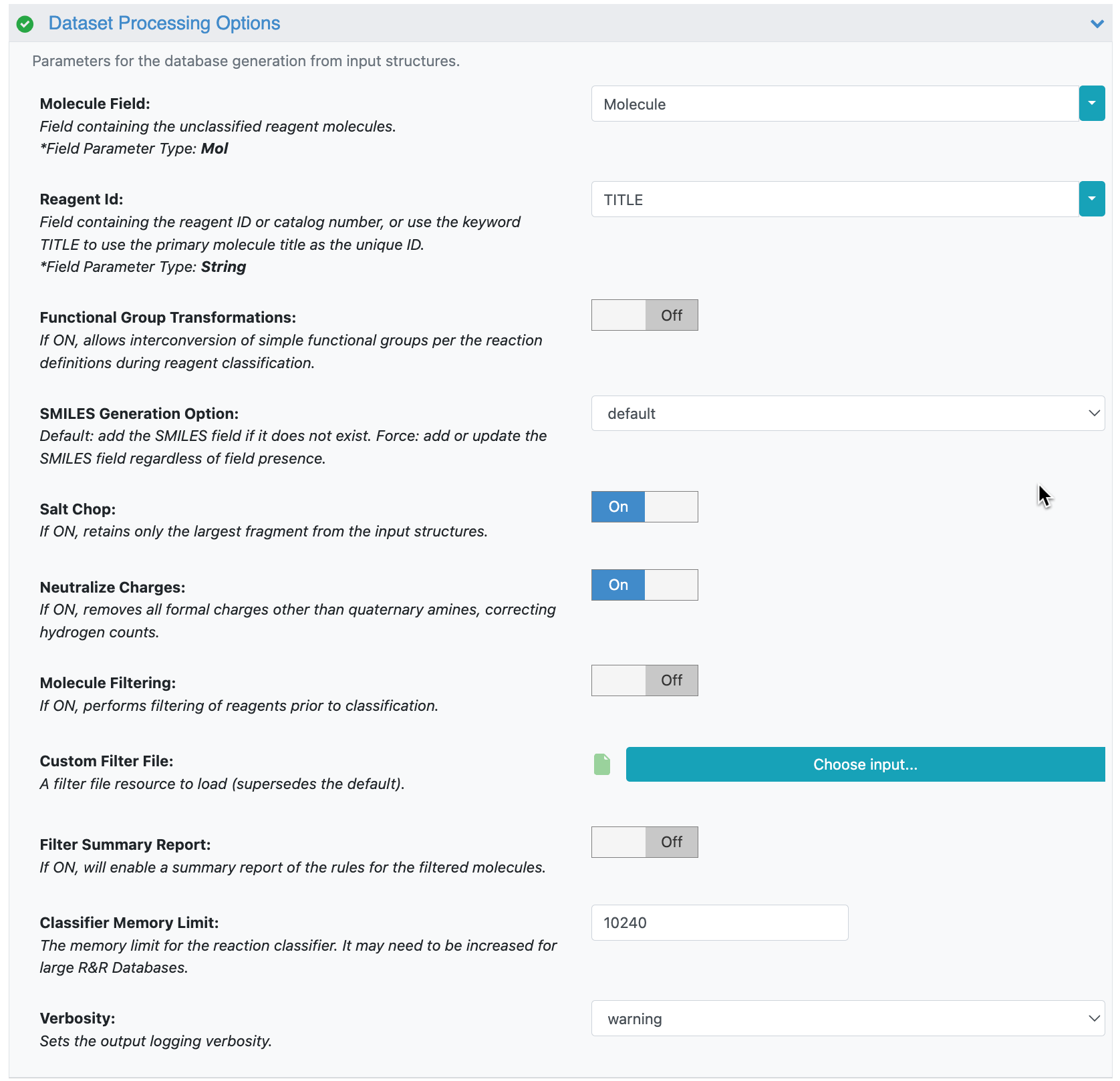
Figure 4. Parameters under the Dataset Processing Options for the Reaction & Reagent Database – Create from Dataset Floe.
Inspect a Reaction and Reagent Database: Writing a Directory
Once generated, a reaction and reagent database can be used as input for any of the reaction-based generative design and enumeration floes. At this point, you have an Orion file resource type. It does not need to be regenerated unless you wish to change the logic or change the reagents contained in it. In this manner, creating the database is “one-stop shopping”: there is no need to go back to chemical structure sources to filter on the basis of functional groups, SMARTS, and so on. This has all been done, for all transformations, by the indexing floe. It is, however, a static file that cannot be easily altered, filtered, and so on. It can be inspected in a couple of different ways. The following tasks will help you to get a sense of what is in the database and to extract reagents that are classified for a particular transformation, which can be used to pull out only reagents of interest into a smaller and more focused database by applying filters (if desired).
Navigate to the Floe page, and launch the Reaction & Reagent Database - Directory Listing Floe.
Select the database file resource that you just wrote. This is the only input required for the floe. Click the “Start Job” button.
To review the results of the floe, navigate to the Jobs Tab and click on the floe you just ran. Select the Floe Report tab to view the generated directory. This shows an exemplar of each reaction in the database and counts the number of each type of reagent that has been classified within the database. The Product Count column shows the number of compounds that would be enumerated if all reagents of each type within the database were allowed to react with one another. Search for the Schotten-Baumann reaction to verify the number of reagents and products in the database. This step is critical before launching the enumeration floe to keep in check the number of products and the size of the collection. The number of products can easily escalate due to the combinatorial nature of the enumeration.
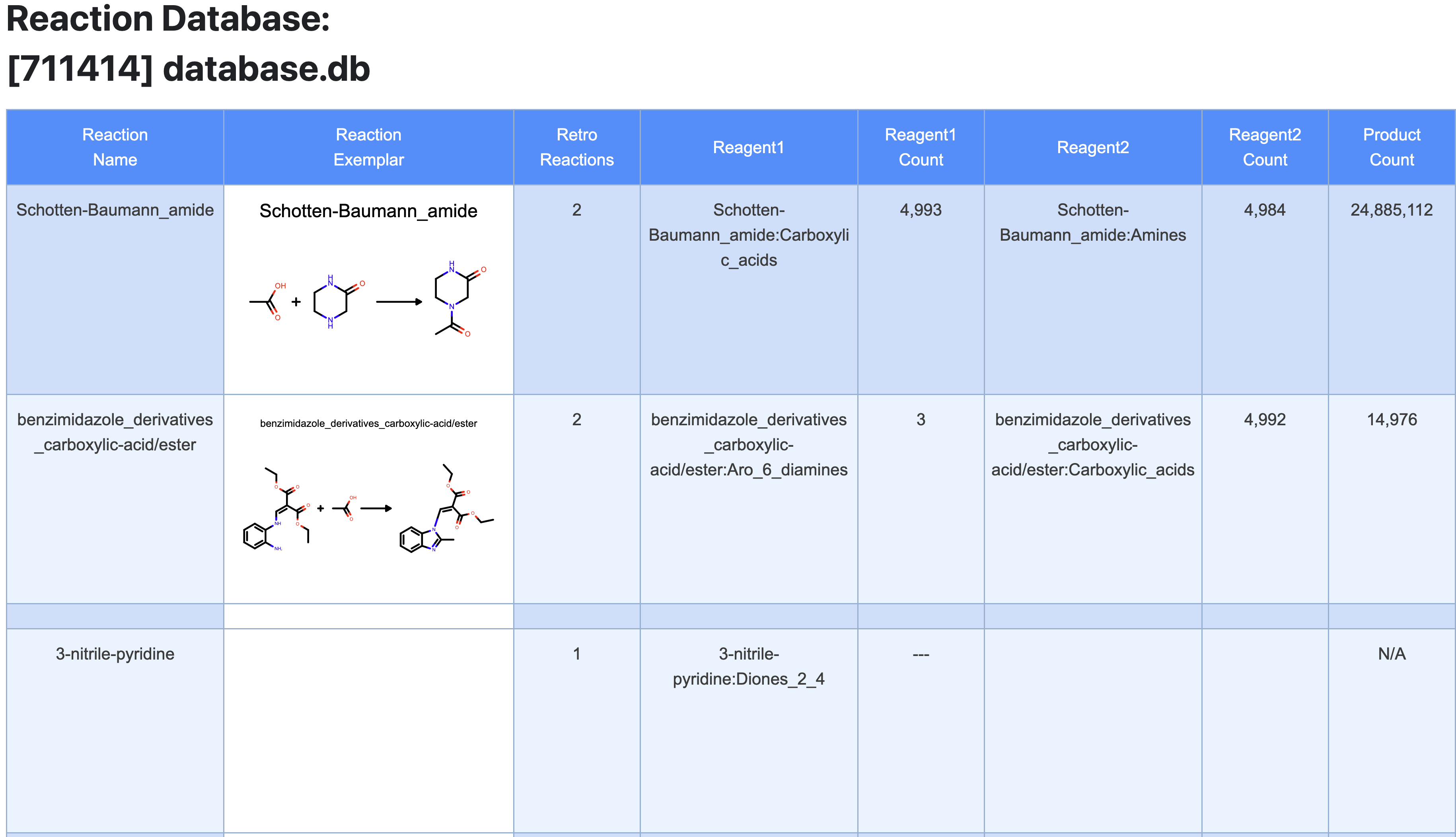
Figure 5. Floe Report from the Reaction & Reagent Database – Directory Listing Floe.
Enumerate Products from a Database with Classified Reagents
The Reaction & Reagent Database - Launch Product Enumerations Floe allows you to enumerate the products of every reaction stored in your database. Alternatively, you can choose to enumerate products from selected reactions, whether built-in or custom. The enumerated products are output to a collection, which can be submitted to downstream hit identification floes for 3D ligand-based or structure-based virtual screening, such as the Prepare Giga Collections Floe. This floe also provides the option to export the products as a SMILES file, which can then be used in 2D ligand-based virtual screening workflows like the Prepare Collection for Fast Similarity or Substructure Search from File Floe. A dataset containing a subset of the products can also be generated, allowing you to inspect the molecules in the 3D & Analyze page or use them as input for OMEGA floes.
Caution
Be mindful when selecting reactions for enumeration. Due to the combinatorial nature of the process, the number of generated products can quickly grow to TRILLIONS of records, which may significantly impact performance, cost, and resource usage.
On the Floe page, select the Reaction & Reagent Database – Launch Product Enumerations Floe and click “Launch Floe” to bring up the Job Form.
Input Parameters
Reaction Database (R&R Enumeration Launcher): Use the database containing the products you intend to enumerate.
Reaction IDs: Remove “All” and select the reaction to be enumerated, Schotten-Baumann_amide in this case. The option “All” enumerates every reaction in the database, which can create a much larger collection with products we are not interested in.
Outputs
Append Enumeration Products to Collection: Give a recognizable name to this output collection.
Product rec/shard: Choose the records per shard for the output collection depending on the floes you intend to run with the output collection. Use 50,000 if this collection will be used with large scale virtual screening floes, such as Gigadock and FastROCS Plus.
Output Product Dataset: Choose a name for the output dataset to capture a subset of the collection. This step is not required, but it allows you to provide a specific name to the output dataset.
Output Product Dataset Limit: Use a limit of 1000. No more than 100,000 should be written to this field, as this is the limit of records that can be examined on the 3D & Analyze page. To learn more about the different file types in Orion, please see a more comprehensive discussion here.
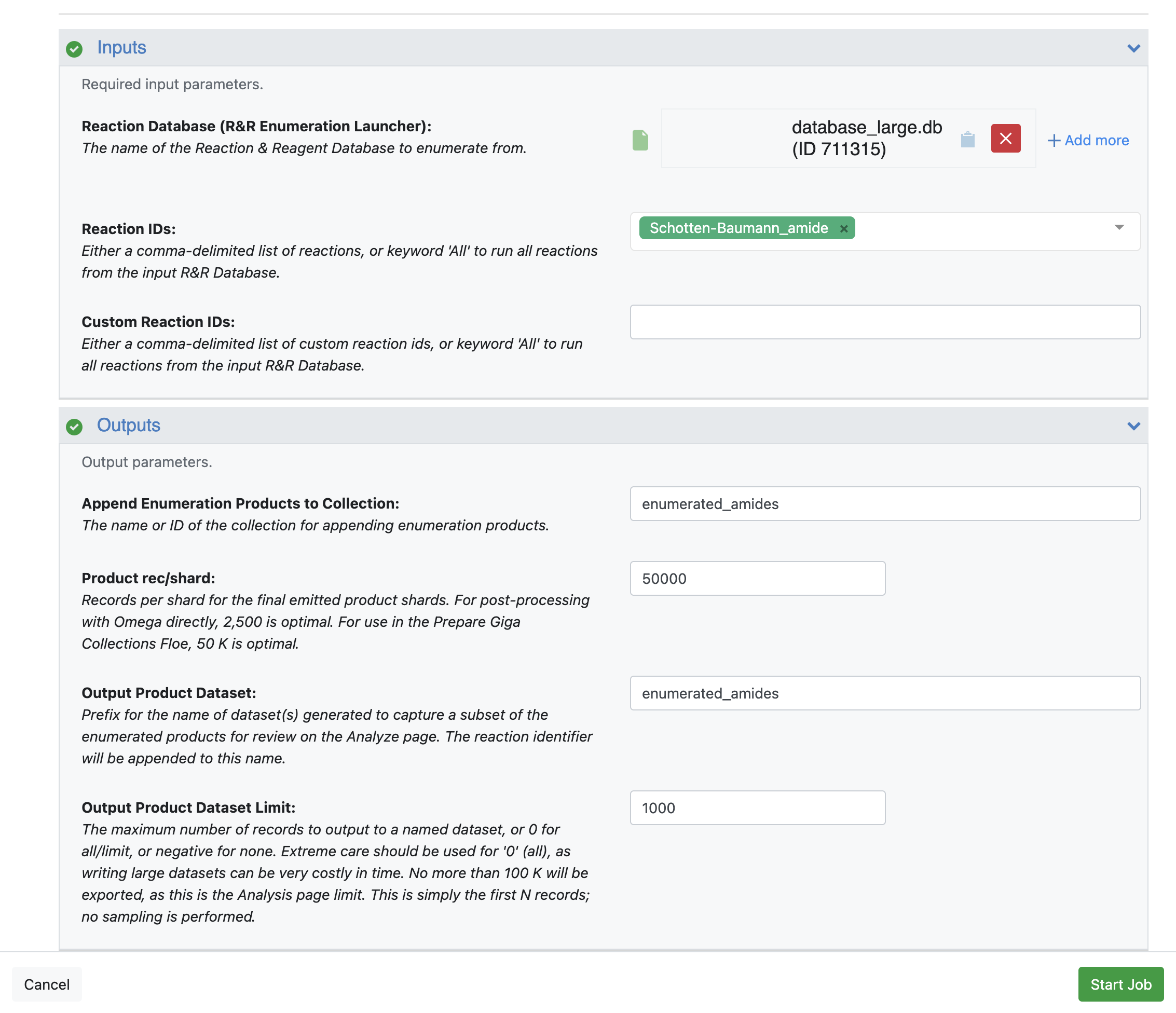
Figure 6. Input and output parameters of the Reaction & Reagent Database – Launch Product Enumerations Floe.
In this tutorial, keep the parameters under the Enumeration Options and Enumeration Run Constraints options at their default values.
Enumeration Advanced Options
Finalization Strategy: Select Force from the drop-down. This option will close the collection once the floe is finished. The default Auto option leaves the collection open in case the user wants to add more products from another enumeration run, which can be achieved by using the same collection name from the Append Enumeration Products to Collection parameter. Please note that the collection size will be displayed as 0 if the Auto option is chosen because it was not closed. The open collection can still be used in downstream floes. In contrast, the Force option closes this collection at the end of the floe.
Output File Resource Name: Choose a name for the generated file that contains the enumerated products and can be exported from Orion.
Output File Format: Select smi from the drop-down.
Click on “Start Job” to launch the enumeration floe. Note that the floe will launch a child job for each reaction enumerated. The main job will give the collection with the enumerated products and a dataset with the job summary. The child jobs will give the output dataset with the subset of the product and the output SMILES file resource.
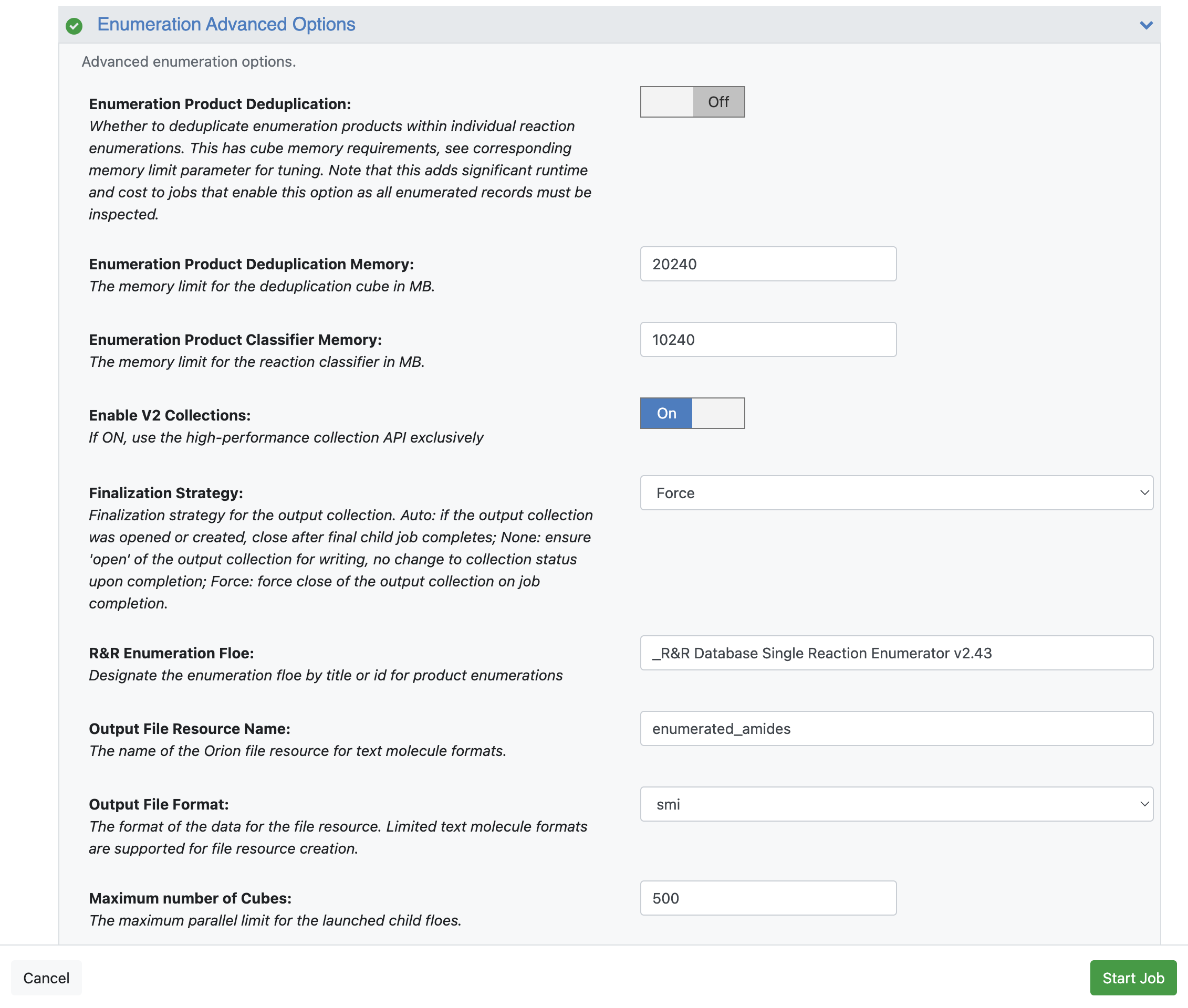
Figure 7. Parameters under the Enumeration Advanced Options for the Reaction & Reagent Database – Launch Product Enumerations Floe.
The output data of the Reaction & Reagent Database – Launch Product Enumerations Floe will be written to the Data page once the floe is completed. A dataset with the job summary will be produced, along with a collection containing all the enumerated products. The child floe, RR_Rxn-Schotten-Baumann_amide, will also generate two outputs: a SMILES file with all enumerated products and a dataset with a subset of the products, which can be inspected and filtered on the 3D & Analyze page.
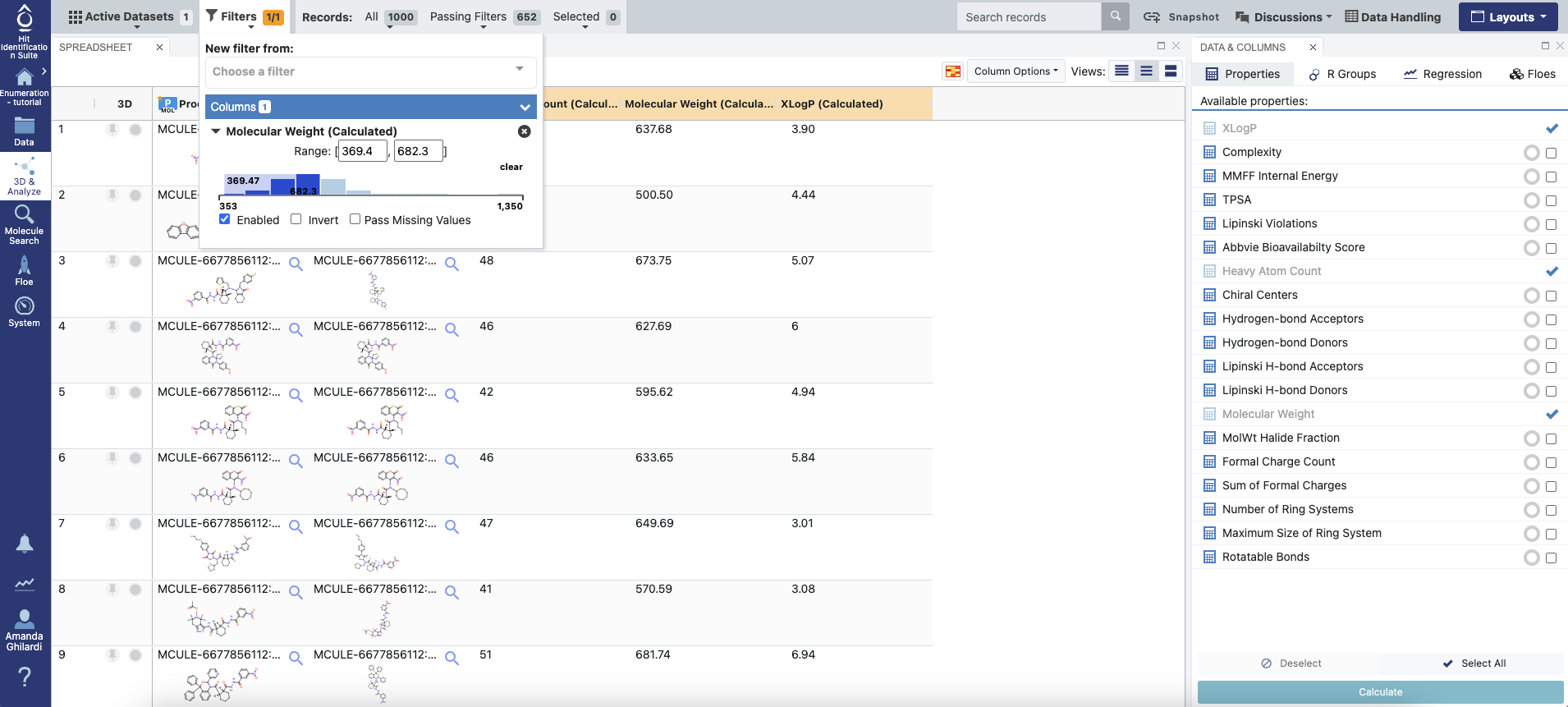
Figure 8. Output dataset of the child floe RR_Rxn-Schotten-Baumann_amide in the 3D & Analyze page. The user can calculate properties from the Available Properties option on the Data & Columns Panel and filter the molecules based on those properties.
Next Steps
The output data from the Reaction & Reagent Database – Launch Product Enumerations Floe can be used directly as input in downstream workflows. The collection containing all the enumerated products can be submitted to the Prepare Giga Collections Floe, which produces ready-to-use collections for the FastROCS and Gigadock workfloes, which are ligand-based and structure-based 3D virtual screening tools, respectively. These floes can be found in the Large-Scale Floes package. In order to use the enumerated products for 2D similarity and substructure search, the SMILES file can be processed using the Prepare Collection for Fast Similarity or Substructure Search from File Floe. This will generate the collections compatible with the following workflows in the ChemInfo Hit ID Floes package: Fast Fingerprint Similarity Search Floe, Fast Substructure Search with SMARTS Floe, and Fast Substructure Search with an MDL Query Floe.
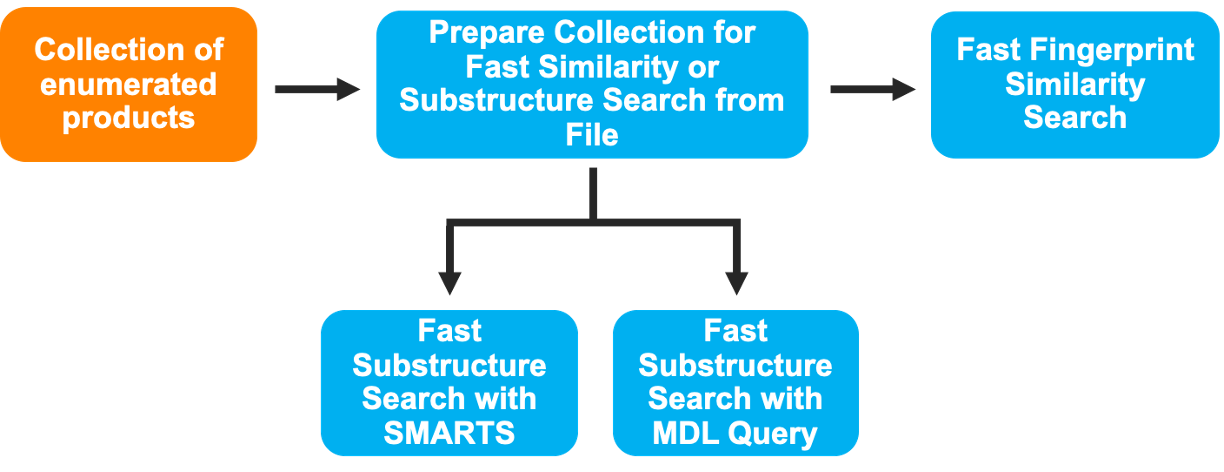
Figure 9. One possible scheme of downstream workflows that can be run using the output data of the Reaction & Reagent Database - Launch Product Enumerations Floe.
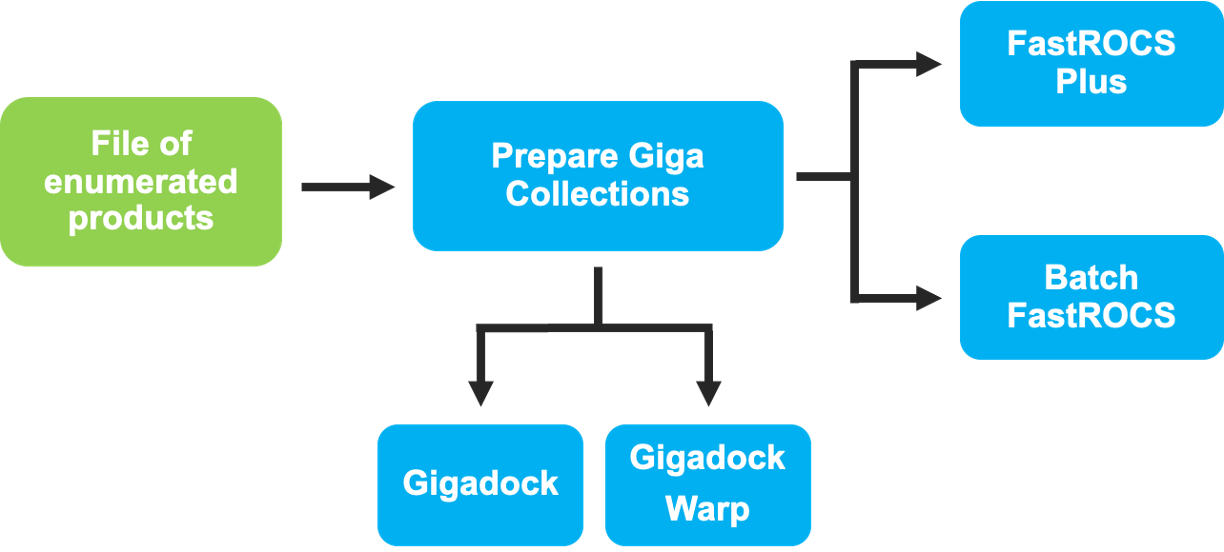
Figure 10. Another scheme of downstream workflows that can be run using the output data of the Reaction & Reagent Database - Launch Product Enumerations Floe.